Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, generate accurate citations for free.
- Knowledge Base
Methodology
- How to Write a Strong Hypothesis | Steps & Examples

How to Write a Strong Hypothesis | Steps & Examples
Published on May 6, 2022 by Shona McCombes . Revised on November 20, 2023.
A hypothesis is a statement that can be tested by scientific research. If you want to test a relationship between two or more variables, you need to write hypotheses before you start your experiment or data collection .
Example: Hypothesis
Daily apple consumption leads to fewer doctor’s visits.
Table of contents
What is a hypothesis, developing a hypothesis (with example), hypothesis examples, other interesting articles, frequently asked questions about writing hypotheses.
A hypothesis states your predictions about what your research will find. It is a tentative answer to your research question that has not yet been tested. For some research projects, you might have to write several hypotheses that address different aspects of your research question.
A hypothesis is not just a guess – it should be based on existing theories and knowledge. It also has to be testable, which means you can support or refute it through scientific research methods (such as experiments, observations and statistical analysis of data).
Variables in hypotheses
Hypotheses propose a relationship between two or more types of variables .
- An independent variable is something the researcher changes or controls.
- A dependent variable is something the researcher observes and measures.
If there are any control variables , extraneous variables , or confounding variables , be sure to jot those down as you go to minimize the chances that research bias will affect your results.
In this example, the independent variable is exposure to the sun – the assumed cause . The dependent variable is the level of happiness – the assumed effect .
Here's why students love Scribbr's proofreading services
Discover proofreading & editing
Step 1. Ask a question
Writing a hypothesis begins with a research question that you want to answer. The question should be focused, specific, and researchable within the constraints of your project.
Step 2. Do some preliminary research
Your initial answer to the question should be based on what is already known about the topic. Look for theories and previous studies to help you form educated assumptions about what your research will find.
At this stage, you might construct a conceptual framework to ensure that you’re embarking on a relevant topic . This can also help you identify which variables you will study and what you think the relationships are between them. Sometimes, you’ll have to operationalize more complex constructs.
Step 3. Formulate your hypothesis
Now you should have some idea of what you expect to find. Write your initial answer to the question in a clear, concise sentence.
4. Refine your hypothesis
You need to make sure your hypothesis is specific and testable. There are various ways of phrasing a hypothesis, but all the terms you use should have clear definitions, and the hypothesis should contain:
- The relevant variables
- The specific group being studied
- The predicted outcome of the experiment or analysis
5. Phrase your hypothesis in three ways
To identify the variables, you can write a simple prediction in if…then form. The first part of the sentence states the independent variable and the second part states the dependent variable.
In academic research, hypotheses are more commonly phrased in terms of correlations or effects, where you directly state the predicted relationship between variables.
If you are comparing two groups, the hypothesis can state what difference you expect to find between them.
6. Write a null hypothesis
If your research involves statistical hypothesis testing , you will also have to write a null hypothesis . The null hypothesis is the default position that there is no association between the variables. The null hypothesis is written as H 0 , while the alternative hypothesis is H 1 or H a .
- H 0 : The number of lectures attended by first-year students has no effect on their final exam scores.
- H 1 : The number of lectures attended by first-year students has a positive effect on their final exam scores.
If you want to know more about the research process , methodology , research bias , or statistics , make sure to check out some of our other articles with explanations and examples.
- Sampling methods
- Simple random sampling
- Stratified sampling
- Cluster sampling
- Likert scales
- Reproducibility
Statistics
- Null hypothesis
- Statistical power
- Probability distribution
- Effect size
- Poisson distribution
Research bias
- Optimism bias
- Cognitive bias
- Implicit bias
- Hawthorne effect
- Anchoring bias
- Explicit bias
A hypothesis is not just a guess — it should be based on existing theories and knowledge. It also has to be testable, which means you can support or refute it through scientific research methods (such as experiments, observations and statistical analysis of data).
Null and alternative hypotheses are used in statistical hypothesis testing . The null hypothesis of a test always predicts no effect or no relationship between variables, while the alternative hypothesis states your research prediction of an effect or relationship.
Hypothesis testing is a formal procedure for investigating our ideas about the world using statistics. It is used by scientists to test specific predictions, called hypotheses , by calculating how likely it is that a pattern or relationship between variables could have arisen by chance.
Cite this Scribbr article
If you want to cite this source, you can copy and paste the citation or click the “Cite this Scribbr article” button to automatically add the citation to our free Citation Generator.
McCombes, S. (2023, November 20). How to Write a Strong Hypothesis | Steps & Examples. Scribbr. Retrieved April 1, 2024, from https://www.scribbr.com/methodology/hypothesis/
Is this article helpful?
Shona McCombes
Other students also liked, construct validity | definition, types, & examples, what is a conceptual framework | tips & examples, operationalization | a guide with examples, pros & cons, what is your plagiarism score.
Loading metrics
Open Access
Perspective
Perspective: Dimensions of the scientific method
* E-mail: [email protected]
Affiliation Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, Georgia, United States of America
- Eberhard O. Voit

Published: September 12, 2019
- https://doi.org/10.1371/journal.pcbi.1007279
- Reader Comments
The scientific method has been guiding biological research for a long time. It not only prescribes the order and types of activities that give a scientific study validity and a stamp of approval but also has substantially shaped how we collectively think about the endeavor of investigating nature. The advent of high-throughput data generation, data mining, and advanced computational modeling has thrown the formerly undisputed, monolithic status of the scientific method into turmoil. On the one hand, the new approaches are clearly successful and expect the same acceptance as the traditional methods, but on the other hand, they replace much of the hypothesis-driven reasoning with inductive argumentation, which philosophers of science consider problematic. Intrigued by the enormous wealth of data and the power of machine learning, some scientists have even argued that significant correlations within datasets could make the entire quest for causation obsolete. Many of these issues have been passionately debated during the past two decades, often with scant agreement. It is proffered here that hypothesis-driven, data-mining–inspired, and “allochthonous” knowledge acquisition, based on mathematical and computational models, are vectors spanning a 3D space of an expanded scientific method. The combination of methods within this space will most certainly shape our thinking about nature, with implications for experimental design, peer review and funding, sharing of result, education, medical diagnostics, and even questions of litigation.
Citation: Voit EO (2019) Perspective: Dimensions of the scientific method. PLoS Comput Biol 15(9): e1007279. https://doi.org/10.1371/journal.pcbi.1007279
Editor: Jason A. Papin, University of Virginia, UNITED STATES
Copyright: © 2019 Eberhard O. Voit. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Funding: This work was supported in part by grants from the National Science Foundation ( https://www.nsf.gov/div/index.jsp?div=MCB ) grant NSF-MCB-1517588 (PI: EOV), NSF-MCB-1615373 (PI: Diana Downs) and the National Institute of Environmental Health Sciences ( https://www.niehs.nih.gov/ ) grant NIH-2P30ES019776-05 (PI: Carmen Marsit). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests: The author has declared that no competing interests exist.
The traditional scientific method: Hypothesis-driven deduction
Research is the undisputed core activity defining science. Without research, the advancement of scientific knowledge would come to a screeching halt. While it is evident that researchers look for new information or insights, the term “research” is somewhat puzzling. Never mind the prefix “re,” which simply means “coming back and doing it again and again,” the word “search” seems to suggest that the research process is somewhat haphazard, that not much of a strategy is involved in the process. One might argue that research a few hundred years ago had the character of hoping for enough luck to find something new. The alchemists come to mind in their quest to turn mercury or lead into gold, or to discover an elixir for eternal youth, through methods we nowadays consider laughable.
Today’s sciences, in stark contrast, are clearly different. Yes, we still try to find something new—and may need a good dose of luck—but the process is anything but unstructured. In fact, it is prescribed in such rigor that it has been given the widely known moniker “scientific method.” This scientific method has deep roots going back to Aristotle and Herophilus (approximately 300 BC), Avicenna and Alhazen (approximately 1,000 AD), Grosseteste and Robert Bacon (approximately 1,250 AD), and many others, but solidified and crystallized into the gold standard of quality research during the 17th and 18th centuries [ 1 – 7 ]. In particular, Sir Francis Bacon (1561–1626) and René Descartes (1596–1650) are often considered the founders of the scientific method, because they insisted on careful, systematic observations of high quality, rather than metaphysical speculations that were en vogue among the scholars of the time [ 1 , 8 ]. In contrast to their peers, they strove for objectivity and insisted that observations, rather than an investigator’s preconceived ideas or superstitions, should be the basis for formulating a research idea [ 7 , 9 ].
Bacon and his 19th century follower John Stuart Mill explicitly proposed gaining knowledge through inductive reasoning: Based on carefully recorded observations, or from data obtained in a well-planned experiment, generalized assertions were to be made about similar yet (so far) unobserved phenomena [ 7 ]. Expressed differently, inductive reasoning attempts to derive general principles or laws directly from empirical evidence [ 10 ]. An example is the 19th century epigram of the physician Rudolf Virchow, Omnis cellula e cellula . There is no proof that indeed “every cell derives from a cell,” but like Virchow, we have made the observation time and again and never encountered anything suggesting otherwise.
In contrast to induction, the widely accepted, traditional scientific method is based on formulating and testing hypotheses. From the results of these tests, a deduction is made whether the hypothesis is presumably true or false. This type of hypotheticodeductive reasoning goes back to William Whewell, William Stanley Jevons, and Charles Peirce in the 19th century [ 1 ]. By the 20th century, the deductive, hypothesis-based scientific method had become deeply ingrained in the scientific psyche, and it is now taught as early as middle school in order to teach students valid means of discovery [ 8 , 11 , 12 ]. The scientific method has not only guided most research studies but also fundamentally influenced how we think about the process of scientific discovery.
Alas, because biology has almost no general laws, deduction in the strictest sense is difficult. It may therefore be preferable to use the term abduction, which refers to the logical inference toward the most plausible explanation, given a set of observations, although this explanation cannot be proven and is not necessarily true.
Over the decades, the hypothesis-based scientific method did experience variations here and there, but its conceptual scaffold remained essentially unchanged ( Fig 1 ). Its key is a process that begins with the formulation of a hypothesis that is to be rigorously tested, either in the wet lab or computationally; nonadherence to this principle is seen as lacking rigor and can lead to irreproducible results [ 1 , 13 – 15 ].
- PPT PowerPoint slide
- PNG larger image
- TIFF original image
The central concept of the traditional scientific method is a falsifiable hypothesis regarding some phenomenon of interest. This hypothesis is to be tested experimentally or computationally. The test results support or refute the hypothesis, triggering a new round of hypothesis formulation and testing.
https://doi.org/10.1371/journal.pcbi.1007279.g001
Going further, the prominent philosopher of science Sir Karl Popper argued that a scientific hypothesis can never be verified but that it can be disproved by a single counterexample. He therefore demanded that scientific hypotheses had to be falsifiable, because otherwise, testing would be moot [ 16 , 17 ] (see also [ 18 ]). As Gillies put it, “successful theories are those that survive elimination through falsification” [ 19 ]. Kelley and Scott agreed to some degree but warned that complete insistence on falsifiability is too restrictive as it would mark many computational techniques, statistical hypothesis testing, and even Darwin’s theory of evolution as nonscientific [ 20 ].
While the hypothesis-based scientific method has been very successful, its exclusive reliance on deductive reasoning is dangerous because according to the so-called Duhem–Quine thesis, hypothesis testing always involves an unknown number of explicit or implicit assumptions, some of which may steer the researcher away from hypotheses that seem implausible, although they are, in fact, true [ 21 ]. According to Kuhn, this bias can obstruct the recognition of paradigm shifts [ 22 ], which require the rethinking of previously accepted “truths” and the development of radically new ideas [ 23 , 24 ]. The testing of simultaneous alternative hypotheses [ 25 – 27 ] ameliorates this problem to some degree but not entirely.
The traditional scientific method is often presented in discrete steps, but it should really be seen as a form of critical thinking, subject to review and independent validation [ 8 ]. It has proven very influential, not only by prescribing valid experimentation, but also for affecting the way we attempt to understand nature [ 18 ], for teaching [ 8 , 12 ], reporting, publishing, and otherwise sharing information [ 28 ], for peer review and the awarding of funds by research-supporting agencies [ 29 , 30 ], for medical diagnostics [ 7 ], and even in litigation [ 31 ].
A second dimension of the scientific method: Data-mining–inspired induction
A major shift in biological experimentation occurred with the–omics revolution of the early 21st century. All of a sudden, it became feasible to perform high-throughput experiments that generated thousands of measurements, typically characterizing the expression or abundances of very many—if not all—genes, proteins, metabolites, or other biological quantities in a sample.
The strategy of measuring large numbers of items in a nontargeted fashion is fundamentally different from the traditional scientific method and constitutes a new, second dimension of the scientific method. Instead of hypothesizing and testing whether gene X is up-regulated under some altered condition, the leading question becomes which of the thousands of genes in a sample are up- or down-regulated. This shift in focus elevates the data to the supreme role of revealing novel insights by themselves ( Fig 2 ). As an important, generic advantage over the traditional strategy, this second dimension is free of a researcher’s preconceived notions regarding the molecular mechanisms governing the phenomenon of interest, which are otherwise the key to formulating a hypothesis. The prominent biologists Patrick Brown and David Botstein commented that “the patterns of expression will often suffice to begin de novo discovery of potential gene functions” [ 32 ].
Data-driven research begins with an untargeted exploration, in which the data speak for themselves. Machine learning extracts patterns from the data, which suggest hypotheses that are to be tested in the lab or computationally.
https://doi.org/10.1371/journal.pcbi.1007279.g002
This data-driven, discovery-generating approach is at once appealing and challenging. On the one hand, very many data are explored simultaneously and essentially without bias. On the other hand, the large datasets supporting this approach create a genuine challenge to understanding and interpreting the experimental results because the thousands of data points, often superimposed with a fair amount of noise, make it difficult to detect meaningful differences between sample and control. This situation can only be addressed with computational methods that first “clean” the data, for instance, through the statistically valid removal of outliers, and then use machine learning to identify statistically significant, distinguishing molecular profiles or signatures. In favorable cases, such signatures point to specific biological pathways, whereas other signatures defy direct explanation but may become the launch pad for follow-up investigations [ 33 ].
Today’s scientists are very familiar with this discovery-driven exploration of “what’s out there” and might consider it a quaint quirk of history that this strategy was at first widely chastised and ridiculed as a “fishing expedition” [ 30 , 34 ]. Strict traditionalists were outraged that rigor was leaving science with the new approach and that sufficient guidelines were unavailable to assure the validity and reproducibility of results [ 10 , 35 , 36 ].
From the view point of philosophy of science, this second dimension of the scientific method uses inductive reasoning and reflects Bacon’s idea that observations can and should dictate the research question to be investigated [ 1 , 7 ]. Allen [ 36 ] forcefully rejected this type of reasoning, stating “the thinking goes, we can now expect computer programs to derive significance, relevance and meaning from chunks of information, be they nucleotide sequences or gene expression profiles… In contrast with this view, many are convinced that no purely logical process can turn observation into understanding.” His conviction goes back to the 18th century philosopher David Hume and again to Popper, who identified as the overriding problem with inductive reasoning that it can never truly reveal causality, even if a phenomenon is observed time and again [ 16 , 17 , 37 , 38 ]. No number of observations, even if they always have the same result, can guard against an exception that would violate the generality of a law inferred from these observations [ 1 , 35 ]. Worse, Popper argued, through inference by induction, we cannot even know the probability of something being true [ 10 , 17 , 36 ].
Others argued that data-driven and hypothesis-driven research actually do not differ all that much in principle, as long as there is cycling between developing new ideas and testing them with care [ 27 ]. In fact, Kell and Oliver [ 34 ] maintained that the exclusive acceptance of hypothesis-driven programs misrepresents the complexities of biological knowledge generation. Similarly refuting the prominent rule of deduction, Platt [ 26 ] and Beard and Kushmerick [ 27 ] argued that repeated inductive reasoning, called strong inference, corresponds to a logically sound decision tree of disproving or refining hypotheses that can rapidly yield firm conclusions; nonetheless, Platt had to admit that inductive inference is not as certain as deduction, because it projects into the unknown. Lander compared the task of obtaining causality by induction to the problem of inferring the design of a microprocessor from input-output readings, which in a strict sense is impossible, because the microprocessor could be arbitrarily complicated; even so, inference often leads to novel insights and therefore is valuable [ 39 ].
An interesting special case of almost pure inductive reasoning is epidemiology, where hypothesis-driven reasoning is rare and instead, the fundamental question is whether data-based evidence is sufficient to associate health risks with specific causes [ 31 , 34 ].
Recent advances in machine learning and “big-data” mining have driven the use of inductive reasoning to unprecedented heights. As an example, machine learning can greatly assist in the discovery of patterns, for instance, in biological sequences [ 40 ]. Going a step further, a pithy article by Andersen [ 41 ] proffered that we may not need to look for causality or mechanistic explanations anymore if we just have enough correlation: “With enough data, the numbers speak for themselves, correlation replaces causation, and science can advance even without coherent models or unified theories.”
Of course, the proposal to abandon the quest for causality caused pushback on philosophical as well as mathematical grounds. Allen [ 10 , 35 ] considered the idea “absurd” that data analysis could enhance understanding in the absence of a hypothesis. He felt confident “that even the formidable combination of computing power with ease of access to data cannot produce a qualitative shift in the way that we do science: the making of hypotheses remains an indispensable component in the growth of knowledge” [ 36 ]. Succi and Coveney [ 42 ] refuted the “most extravagant claims” of big-data proponents very differently, namely by analyzing the theories on which machine learning is founded. They contrasted the assumptions underlying these theories, such as the law of large numbers, with the mathematical reality of complex biological systems. Specifically, they carefully identified genuine features of these systems, such as nonlinearities, nonlocality of effects, fractal aspects, and high dimensionality, and argued that they fundamentally violate some of the statistical assumptions implicitly underlying big-data analysis, like independence of events. They concluded that these discrepancies “may lead to false expectations and, at their nadir, even to dangerous social, economical and political manipulation.” To ameliorate the situation, the field of big-data analysis would need new strong theorems characterizing the validity of its methods and the numbers of data required for obtaining reliable insights. Succi and Coveney go as far as stating that too many data are just as bad as insufficient data [ 42 ].
While philosophical doubts regarding inductive methods will always persist, one cannot deny that -omics-based, high-throughput studies, combined with machine learning and big-data analysis, have been very successful [ 43 ]. Yes, induction cannot truly reveal general laws, no matter how large the datasets, but they do provide insights that are very different from what science had offered before and may at least suggest novel patterns, trends, or principles. As a case in point, if many transcriptomic studies indicate that a particular gene set is involved in certain classes of phenomena, there is probably some truth to the observation, even though it is not mathematically provable. Kepler’s laws of astronomy were arguably derived solely from inductive reasoning [ 34 ].
Notwithstanding the opposing views on inductive methods, successful strategies shape how we think about science. Thus, to take advantage of all experimental options while ensuring quality of research, we must not allow that “anything goes” but instead identify and characterize standard operating procedures and controls that render this emerging scientific method valid and reproducible. A laudable step in this direction was the wide acceptance of “minimum information about a microarray experiment” (MIAME) standards for microarray experiments [ 44 ].
A third dimension of the scientific method: Allochthonous reasoning
Parallel to the blossoming of molecular biology and the rapid rise in the power and availability of computing in the late 20th century, the use of mathematical and computational models became increasingly recognized as relevant and beneficial for understanding biological phenomena. Indeed, mathematical models eventually achieved cornerstone status in the new field of computational systems biology.
Mathematical modeling has been used as a tool of biological analysis for a long time [ 27 , 45 – 48 ]. Interesting for the discussion here is that the use of mathematical and computational modeling in biology follows a scientific approach that is distinctly different from the traditional and the data-driven methods, because it is distributed over two entirely separate domains of knowledge. One consists of the biological reality of DNA, elephants, and roses, whereas the other is the world of mathematics, which is governed by numbers, symbols, theorems, and abstract work protocols. Because the ways of thinking—and even the languages—are different in these two realms, I suggest calling this type of knowledge acquisition “allochthonous” (literally Greek: in or from a “piece of land different from where one is at home”; one could perhaps translate it into modern lingo as “outside one’s comfort zone”). De facto, most allochthonous reasoning in biology presently refers to mathematics and computing, but one might also consider, for instance, the application of methods from linguistics in the analysis of DNA sequences or proteins [ 49 ].
One could argue that biologists have employed “models” for a long time, for instance, in the form of “model organisms,” cell lines, or in vitro experiments, which more or less faithfully reflect features of the organisms of true interest but are easier to manipulate. However, this type of biological model use is rather different from allochthonous reasoning, as it does not leave the realm of biology and uses the same language and often similar methodologies.
A brief discussion of three experiences from our lab may illustrate the benefits of allochthonous reasoning. (1) In a case study of renal cell carcinoma, a dynamic model was able to explain an observed yet nonintuitive metabolic profile in terms of the enzymatic reaction steps that had been altered during the disease [ 50 ]. (2) A transcriptome analysis had identified several genes as displaying significantly different expression patterns during malaria infection in comparison to the state of health. Considered by themselves and focusing solely on genes coding for specific enzymes of purine metabolism, the findings showed patterns that did not make sense. However, integrating the changes in a dynamic model revealed that purine metabolism globally shifted, in response to malaria, from guanine compounds to adenine, inosine, and hypoxanthine [ 51 ]. (3) Data capturing the dynamics of malaria parasites suggested growth rates that were biologically impossible. Speculation regarding possible explanations led to the hypothesis that many parasite-harboring red blood cells might “hide” from circulation and therewith from detection in the blood stream. While experimental testing of the feasibility of the hypothesis would have been expensive, a dynamic model confirmed that such a concealment mechanism could indeed quantitatively explain the apparently very high growth rates [ 52 ]. In all three cases, the insights gained inductively from computational modeling would have been difficult to obtain purely with experimental laboratory methods. Purely deductive allochthonous reasoning is the ultimate goal of the search for design and operating principles [ 53 – 55 ], which strives to explain why certain structures or functions are employed by nature time and again. An example is a linear metabolic pathway, in which feedback inhibition is essentially always exerted on the first step [ 56 , 57 ]. This generality allows the deduction that a so far unstudied linear pathway is most likely (or even certain to be) inhibited at the first step. Not strictly deductive—but rather abductive—was a study in our lab in which we analyzed time series data with a mathematical model that allowed us to infer the most likely regulatory structure of a metabolic pathway [ 58 , 59 ].
A typical allochthonous investigation begins in the realm of biology with the formulation of a hypothesis ( Fig 3 ). Instead of testing this hypothesis with laboratory experiments, the system encompassing the hypothesis is moved into the realm of mathematics. This move requires two sets of ingredients. One set consists of the simplification and abstraction of the biological system: Any distracting details that seem unrelated to the hypothesis and its context are omitted or represented collectively with other details. This simplification step carries the greatest risk of the entire modeling approach, as omission of seemingly negligible but, in truth, important details can easily lead to wrong results. The second set of ingredients consists of correspondence rules that translate every biological component or process into the language of mathematics [ 60 , 61 ].
This mathematical and computational approach is distributed over two realms, which are connected by correspondence rules.
https://doi.org/10.1371/journal.pcbi.1007279.g003
Once the system is translated, it has become an entirely mathematical construct that can be analyzed purely with mathematical and computational means. The results of this analysis are also strictly mathematical. They typically consist of values of variables, magnitudes of processes, sensitivity patterns, signs of eigenvalues, or qualitative features like the onset of oscillations or the potential for limit cycles. Correspondence rules are used again to move these results back into the realm of biology. As an example, the mathematical result that “two eigenvalues have positive real parts” does not make much sense to many biologists, whereas the interpretation that “the system is not stable at the steady state in question” is readily explained. New biological insights may lead to new hypotheses, which are tested either by experiments or by returning once more to the realm of mathematics. The model design, diagnosis, refinements, and validation consist of several phases, which have been discussed widely in the biomathematical literature. Importantly, each iteration of a typical modeling analysis consists of a move from the biological to the mathematical realm and back.
The reasoning within the realm of mathematics is often deductive, in the form of an Aristotelian syllogism, such as the well-known “All men are mortal; Socrates is a man; therefore, Socrates is mortal.” However, the reasoning may also be inductive, as it is the case with large-scale Monte-Carlo simulations that generate arbitrarily many “observations,” although they cannot reveal universal principles or theorems. An example is a simulation randomly drawing numbers in an attempt to show that every real number has an inverse. The simulation will always attest to this hypothesis but fail to discover the truth because it will never randomly draw 0. Generically, computational models may be considered sets of hypotheses, formulated as equations or as algorithms that reflect our perception of a complex system [ 27 ].
Impact of the multidimensional scientific method on learning
Almost all we know in biology has come from observation, experimentation, and interpretation. The traditional scientific method not only offered clear guidance for this knowledge gathering, but it also fundamentally shaped the way we think about the exploration of nature. When presented with a new research question, scientists were trained to think immediately in terms of hypotheses and alternatives, pondering the best feasible ways of testing them, and designing in their minds strong controls that would limit the effects of known or unknown confounders. Shaped by the rigidity of this ever-repeating process, our thinking became trained to move forward one well-planned step at a time. This modus operandi was rigid and exact. It also minimized the erroneous pursuit of long speculative lines of thought, because every step required testing before a new hypothesis was formed. While effective, the process was also very slow and driven by ingenuity—as well as bias—on the scientist’s part. This bias was sometimes a hindrance to necessary paradigm shifts [ 22 ].
High-throughput data generation, big-data analysis, and mathematical-computational modeling changed all that within a few decades. In particular, the acceptance of inductive principles and of the allochthonous use of nonbiological strategies to answer biological questions created an unprecedented mix of successes and chaos. To the horror of traditionalists, the importance of hypotheses became minimized, and the suggestion spread that the data would speak for themselves [ 36 ]. Importantly, within this fog of “anything goes,” the fundamental question arose how to determine whether an experiment was valid.
Because agreed-upon operating procedures affect research progress and interpretation, thinking, teaching, and sharing of results, this question requires a deconvolution of scientific strategies. Here I proffer that the single scientific method of the past should be expanded toward a vector space of scientific methods, with spanning vectors that correspond to different dimensions of the scientific method ( Fig 4 ).
The traditional hypothesis-based deductive scientific method is expanded into a 3D space that allows for synergistic blends of methods that include data-mining–inspired, inductive knowledge acquisition, and mathematical model-based, allochthonous reasoning.
https://doi.org/10.1371/journal.pcbi.1007279.g004
Obviously, all three dimensions have their advantages and drawbacks. The traditional, hypothesis-driven deductive method is philosophically “clean,” except that it is confounded by preconceptions and assumptions. The data-mining–inspired inductive method cannot offer universal truths but helps us explore very large spaces of factors that contribute to a phenomenon. Allochthonous, model-based reasoning can be performed mentally, with paper and pencil, through rigorous analysis, or with a host of computational methods that are precise and disprovable [ 27 ]. At the same time, they are incomparable faster, cheaper, and much more comprehensive than experiments in molecular biology. This reduction in cost and time, and the increase in coverage, may eventually have far-reaching consequences, as we can already fathom from much of modern physics.
Due to its long history, the traditional dimension of the scientific method is supported by clear and very strong standard operating procedures. Similarly, strong procedures need to be developed for the other two dimensions. The MIAME rules for microarray analysis provide an excellent example [ 44 ]. On the mathematical modeling front, no such rules are generally accepted yet, but trends toward them seem to emerge at the horizon. For instance, it seems to be becoming common practice to include sensitivity analyses in typical modeling studies and to assess the identifiability or sloppiness of ensembles of parameter combinations that fit a given dataset well [ 62 , 63 ].
From a philosophical point of view, it seems unlikely that objections against inductive reasoning will disappear. However, instead of pitting hypothesis-based deductive reasoning against inductivism, it seems more beneficial to determine how the different methods can be synergistically blended ( cf . [ 18 , 27 , 34 , 42 ]) as linear combinations of the three vectors of knowledge acquisition ( Fig 4 ). It is at this point unclear to what degree the identified three dimensions are truly independent of each other, whether additional dimensions should be added [ 24 ], or whether the different versions could be amalgamated into a single scientific method [ 18 ], especially if it is loosely defined as a form of critical thinking [ 8 ]. Nobel Laureate Percy Bridgman even concluded that “science is what scientists do, and there are as many scientific methods as there are individual scientists” [ 8 , 64 ].
Combinations of the three spanning vectors of the scientific method have been emerging for some time. Many biologists already use inductive high-throughput methods to develop specific hypotheses that are subsequently tested with deductive or further inductive methods [ 34 , 65 ]. In terms of including mathematical modeling, physics and geology have been leading the way for a long time, often by beginning an investigation in theory, before any actual experiment is performed. It will benefit biology to look into this strategy and to develop best practices of allochthonous reasoning.
The blending of methods may take quite different shapes. Early on, Ideker and colleagues [ 65 ] proposed an integrated experimental approach for pathway analysis that offered a glimpse of new experimental strategies within the space of scientific methods. In a similar vein, Covert and colleagues [ 66 ] included computational methods into such an integrated approach. Additional examples of blended analyses in systems biology can be seen in other works, such as [ 43 , 67 – 73 ]. Generically, it is often beneficial to start with big data, determine patterns in associations and correlations, then switch to the mathematical realm in order to filter out spurious correlations in a high-throughput fashion. If this procedure is executed in an iterative manner, the “surviving” associations have an increased level of confidence and are good candidates for further experimental or computational testing (personal communication from S. Chandrasekaran).
If each component of a blended scientific method follows strict, commonly agreed guidelines, “linear combinations” within the 3D space can also be checked objectively, per deconvolution. In addition, guidelines for synergistic blends of component procedures should be developed. If we carefully monitor such blends, time will presumably indicate which method is best for which task and how the different approaches optimally inform each other. For instance, it will be interesting to study whether there is an optimal sequence of experiments along the three axes for a particular class of tasks. Big-data analysis together with inductive reasoning might be optimal for creating initial hypotheses and possibly refuting wrong speculations (“we had thought this gene would be involved, but apparently it isn’t”). If the logic of an emerging hypotheses can be tested with mathematical and computational tools, it will almost certainly be faster and cheaper than an immediate launch into wet-lab experimentation. It is also likely that mathematical reasoning will be able to refute some apparently feasible hypothesis and suggest amendments. Ultimately, the “surviving” hypotheses must still be tested for validity through conventional experiments. Deconvolving current practices and optimizing the combination of methods within the 3D or higher-dimensional space of scientific methods will likely result in better planning of experiments and in synergistic blends of approaches that have the potential capacity of addressing some of the grand challenges in biology.
Acknowledgments
The author is very grateful to Dr. Sriram Chandrasekaran and Ms. Carla Kumbale for superb suggestions and invaluable feedback.
- View Article
- Google Scholar
- 2. Gauch HGJ. Scientific Method in Brief. Cambridge, UK.: Cambridge University Press; 2012.
- 3. Gimbel S (Ed). Exploring the Scientific Method: Cases and Questions. Chicago, IL: The University of Chicago Press; 2011.
- PubMed/NCBI
- 8. McLelland CV. The nature of science and the scientific method. Boulder, CO: The Geological Society of America; 2006.
- 9. Ladyman J. Understanding Philosophy of Science. Abington, Oxon: Routledge; 2002.
- 16. Popper KR. Conjectures and Refutations: The Growth of Scientific Knowledge. Abingdon, Oxon: Routledge and Kegan Paul; 1963.
- 17. Popper KR. The Logic of Scientific Discovery. Abingdon, Oxon: Routledge; 2002.
- 21. Harding SE. Can theories be refuted?: Essays on the Duhem-Quine thesis. Dordrecht-Holland / Boston, MA: D. Reidel Publ. Co; 1976.
- 22. Kuhn TS. The Structure of Scientific Revolutions. Chicago, IL: University of Chicago Press; 1962.
- 37. Hume D. An enquiry concerning human understanding. Oxford, U.K.: Oxford University Press; 1748/1999.
- 38. Popper KR. Objective knowledge. An evolutionary approach. Oxford, U.K.: Oxford University Press; 1972.
- 47. von Bertalanffy L. General System Theory: Foundations, Development, Applications. New York: George Braziller; 1968.
- 48. May RM. Stability and Complexity in Model Ecosystems: Princeton University Press; 1973.
- 57. Savageau MA. Biochemical Systems Analysis: A Study of Function and Design in Molecular Biology. Reading, Mass: Addison-Wesley Pub. Co. Advanced Book Program (reprinted 2009); 1976.
- 60. Reither F. Über das Denken mit Analogien und Modellen. In: Schaefer G, Trommer G, editors. Denken in Modellen. Braunschweig, Germany: Georg Westermann Verlag; 1977.
- 61. Voit EO. A First Course in Systems Biology, 2nd Ed. New York, NY: Garland Science; 2018.
- 64. Bridgman PW. Reflections of a Physicist. New York, NY: reprinted by Kessinger Legacy Reprints, 2010; 1955.

How to Implement Hypothesis-Driven Development
Remember back to the time when we were in high school science class. Our teachers had a framework for helping us learn – an experimental approach based on the best available evidence at hand. We were asked to make observations about the world around us, then attempt to form an explanation or hypothesis to explain what we had observed. We then tested this hypothesis by predicting an outcome based on our theory that would be achieved in a controlled experiment – if the outcome was achieved, we had proven our theory to be correct.
We could then apply this learning to inform and test other hypotheses by constructing more sophisticated experiments, and tuning, evolving or abandoning any hypothesis as we made further observations from the results we achieved.
Experimentation is the foundation of the scientific method, which is a systematic means of exploring the world around us. Although some experiments take place in laboratories, it is possible to perform an experiment anywhere, at any time, even in software development.
Practicing Hypothesis-Driven Development is thinking about the development of new ideas, products and services – even organizational change – as a series of experiments to determine whether an expected outcome will be achieved. The process is iterated upon until a desirable outcome is obtained or the idea is determined to be not viable.
We need to change our mindset to view our proposed solution to a problem statement as a hypothesis, especially in new product or service development – the market we are targeting, how a business model will work, how code will execute and even how the customer will use it.
We do not do projects anymore, only experiments. Customer discovery and Lean Startup strategies are designed to test assumptions about customers. Quality Assurance is testing system behavior against defined specifications. The experimental principle also applies in Test-Driven Development – we write the test first, then use the test to validate that our code is correct, and succeed if the code passes the test. Ultimately, product or service development is a process to test a hypothesis about system behaviour in the environment or market it is developed for.
The key outcome of an experimental approach is measurable evidence and learning.
Learning is the information we have gained from conducting the experiment. Did what we expect to occur actually happen? If not, what did and how does that inform what we should do next?
In order to learn we need use the scientific method for investigating phenomena, acquiring new knowledge, and correcting and integrating previous knowledge back into our thinking.
As the software development industry continues to mature, we now have an opportunity to leverage improved capabilities such as Continuous Design and Delivery to maximize our potential to learn quickly what works and what does not. By taking an experimental approach to information discovery, we can more rapidly test our solutions against the problems we have identified in the products or services we are attempting to build. With the goal to optimize our effectiveness of solving the right problems, over simply becoming a feature factory by continually building solutions.
The steps of the scientific method are to:
- Make observations
- Formulate a hypothesis
- Design an experiment to test the hypothesis
- State the indicators to evaluate if the experiment has succeeded
- Conduct the experiment
- Evaluate the results of the experiment
- Accept or reject the hypothesis
- If necessary, make and test a new hypothesis
Using an experimentation approach to software development
We need to challenge the concept of having fixed requirements for a product or service. Requirements are valuable when teams execute a well known or understood phase of an initiative, and can leverage well understood practices to achieve the outcome. However, when you are in an exploratory, complex and uncertain phase you need hypotheses.
Handing teams a set of business requirements reinforces an order-taking approach and mindset that is flawed.
Business does the thinking and ‘knows’ what is right. The purpose of the development team is to implement what they are told. But when operating in an area of uncertainty and complexity, all the members of the development team should be encouraged to think and share insights on the problem and potential solutions. A team simply taking orders from a business owner is not utilizing the full potential, experience and competency that a cross-functional multi-disciplined team offers.
Framing hypotheses
The traditional user story framework is focused on capturing requirements for what we want to build and for whom, to enable the user to receive a specific benefit from the system.
As A…. <role>
I Want… <goal/desire>
So That… <receive benefit>
Behaviour Driven Development (BDD) and Feature Injection aims to improve the original framework by supporting communication and collaboration between developers, tester and non-technical participants in a software project.
In Order To… <receive benefit>
As A… <role>
When viewing work as an experiment, the traditional story framework is insufficient. As in our high school science experiment, we need to define the steps we will take to achieve the desired outcome. We then need to state the specific indicators (or signals) we expect to observe that provide evidence that our hypothesis is valid. These need to be stated before conducting the test to reduce biased interpretations of the results.
If we observe signals that indicate our hypothesis is correct, we can be more confident that we are on the right path and can alter the user story framework to reflect this.
Therefore, a user story structure to support Hypothesis-Driven Development would be;
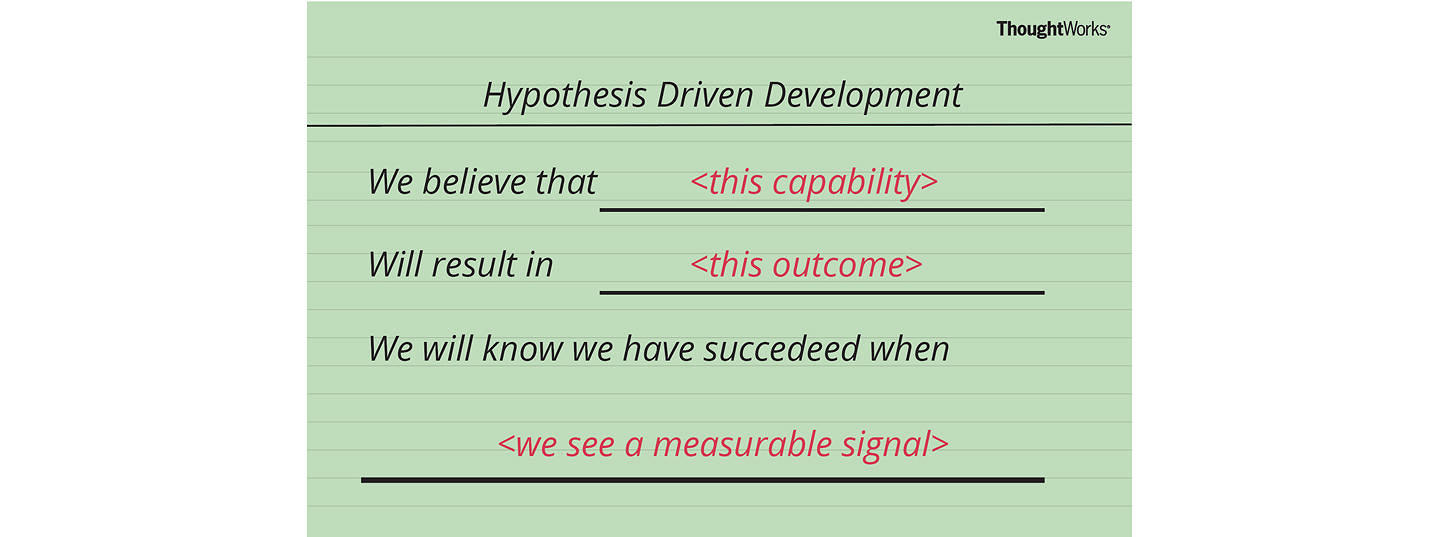
We believe < this capability >
What functionality we will develop to test our hypothesis? By defining a ‘test’ capability of the product or service that we are attempting to build, we identify the functionality and hypothesis we want to test.
Will result in < this outcome >
What is the expected outcome of our experiment? What is the specific result we expect to achieve by building the ‘test’ capability?
We will know we have succeeded when < we see a measurable signal >
What signals will indicate that the capability we have built is effective? What key metrics (qualitative or quantitative) we will measure to provide evidence that our experiment has succeeded and give us enough confidence to move to the next stage.
The threshold you use for statistically significance will depend on your understanding of the business and context you are operating within. Not every company has the user sample size of Amazon or Google to run statistically significant experiments in a short period of time. Limits and controls need to be defined by your organization to determine acceptable evidence thresholds that will allow the team to advance to the next step.
For example if you are building a rocket ship you may want your experiments to have a high threshold for statistical significance. If you are deciding between two different flows intended to help increase user sign up you may be happy to tolerate a lower significance threshold.
The final step is to clearly and visibly state any assumptions made about our hypothesis, to create a feedback loop for the team to provide further input, debate and understanding of the circumstance under which we are performing the test. Are they valid and make sense from a technical and business perspective?
Hypotheses when aligned to your MVP can provide a testing mechanism for your product or service vision. They can test the most uncertain areas of your product or service, in order to gain information and improve confidence.
Examples of Hypothesis-Driven Development user stories are;
Business story
We Believe That increasing the size of hotel images on the booking page
Will Result In improved customer engagement and conversion
We Will Know We Have Succeeded When we see a 5% increase in customers who review hotel images who then proceed to book in 48 hours.
It is imperative to have effective monitoring and evaluation tools in place when using an experimental approach to software development in order to measure the impact of our efforts and provide a feedback loop to the team. Otherwise we are essentially blind to the outcomes of our efforts.
In agile software development we define working software as the primary measure of progress.
By combining Continuous Delivery and Hypothesis-Driven Development we can now define working software and validated learning as the primary measures of progress.
Ideally we should not say we are done until we have measured the value of what is being delivered – in other words, gathered data to validate our hypothesis.
Examples of how to gather data is performing A/B Testing to test a hypothesis and measure to change in customer behaviour. Alternative testings options can be customer surveys, paper prototypes, user and/or guerrilla testing.
One example of a company we have worked with that uses Hypothesis-Driven Development is lastminute.com . The team formulated a hypothesis that customers are only willing to pay a max price for a hotel based on the time of day they book. Tom Klein, CEO and President of Sabre Holdings shared the story of how they improved conversion by 400% within a week.
Combining practices such as Hypothesis-Driven Development and Continuous Delivery accelerates experimentation and amplifies validated learning. This gives us the opportunity to accelerate the rate at which we innovate while relentlessly reducing cost, leaving our competitors in the dust. Ideally we can achieve the ideal of one piece flow: atomic changes that enable us to identify causal relationships between the changes we make to our products and services, and their impact on key metrics.
As Kent Beck said, “Test-Driven Development is a great excuse to think about the problem before you think about the solution”. Hypothesis-Driven Development is a great opportunity to test what you think the problem is, before you work on the solution.
How can you achieve faster growth?
Hypothesis-driven science in large-scale studies: the case of GWAS
- Open access
- Published: 19 September 2021
- Volume 36 , article number 46 , ( 2021 )
Cite this article
You have full access to this open access article
- James Read ORCID: orcid.org/0000-0003-2226-0340 1 &
- Sumana Sharma ORCID: orcid.org/0000-0003-0598-2181 2
6528 Accesses
3 Citations
1 Altmetric
Explore all metrics
It is now well-appreciated by philosophers that contemporary large-scale ‘-omics’ studies in biology stand in non-trivial relationships to more orthodox hypothesis-driven approaches. These relationships have been clarified by Ratti ( 2015 ); however, there remains much more to be said regarding how an important field of genomics cited in that work—‘genome-wide association studies’ (GWAS)—fits into this framework. In the present article, we propose a revision to Ratti’s framework more suited to studies such as GWAS. In the process of doing so, we introduce to the philosophical literature novel exploratory experiments in (phospho)proteomics, and demonstrate how these experiments interplay with the above considerations.
Similar content being viewed by others
Raiders of the lost HARK: a reproducible inference framework for big data science
Mattia Prosperi, Jiang Bian, … Mo Wang

On Fishing for Significance and Statistician’s Degree of Freedom in the Era of Big Molecular Data

Experimental Discovery, Data Models, and Mechanisms in Biology: An Example from Mendel’s Work
Avoid common mistakes on your manuscript.
Introduction
The fields of molecular biology and genetics were transformed upon completion in 2001 of the Human Genome Project (Lander et al. 2001 ). This provided for the first time near-complete information on the genetic makeup of human beings, and marked the advent of what has become known as the ‘post-genomics’ era, defined by the availability of large-scale data sets derived from ‘genome-scale’ approaches. In turn, this has led to a shift in biological methodology, from carefully constructed hypothesis-driven research, to unbiased data-driven approaches, sometimes called ‘-omics’ studies. These studies have attracted philosophical interest in recent years: see e.g. Burian ( 2007 ); O’Malley et al. ( 2010 ); Ratti ( 2015 ); for more general philosophical discussions of large-scale data-driven approaches in contemporary post-genomics biology, see e.g. Leonelli ( 2016 ); Richardson and Stevens ( 2015 ).
Recall that -omics studies fall into three main categories: ‘genomics’, ‘transcriptomics’, and ‘proteomics’. The salient features of these three categories as as follows (we make no claim that these features exhaust any of the three categories; they are, however, the features which are relevant to the present article). Genomics is the study of the complete set of genes (composed of DNA) inside a cell. Cellular processes lead to genetic information being transcribed (copied) into molecules known as RNA. ‘Messenger RNA’ (mRNA) carries information corresponding to the genetic sequence of a gene. Transcriptomics is the study of the complete set of RNA transcripts that are produced by the genome. Finally, the information encoded in mRNA is used by cellular machinery called ribosomes to construct proteins; proteomics is the systematic study of these proteins within a cell. Proteins are the ultimate workhorses of the cell; proteomics studies aim to characterise cellular functions mediated by protein networks, in which nodes represent proteins and edges represent physical/functional interactions between them. For further background on genomics, transcriptomics, and proteomics, see Hasin et al. ( 2017 ).
Large-scale -omics studies are often described as being ‘hypothesis-free’. To take one example from genomics: advances in genome-editing techniques mean that it is now possible to generate ‘loss-of-function’ mutants in the laboratory. Such mutations are inactivating in the sense that they lead to the loss in the function of a gene within a cell. In the last few years, CRISPR-Cas9 technology has emerged, which makes it possible to create targetted loss-of-function mutants for any of the nearly 20,000 genes in the human genome (Doudna and Charpentier 2014 ). This allows researchers to ‘screen’ for a gene the loss of which leads to the phenotype of interest, thereby identifying the function of that gene. The methodological idea behind such screening approaches is that one does not require any background hypothesis as to which gene could be involved in a particular biological process, or associated with a particular phenotype: hence the widespread declaration that such approaches are ‘hypothesis-free’ (Shalem et al. 2015 ). As Burian writes, “Genomics, proteomics, and related “omics” disciplines represent a break with the ideal of hypothesis-driven science” (Burian 2007 , p. 289).
With Ratti ( 2015 ); Franklin ( 2005 ), and others, we find the terminology of ‘hypothesis-free’ to be misleading—for, in fact, such large-scale studies exhibit a Janus-faced dependence on mechanistic hypotheses of a quite standard sort. Ratti characterises such studies, and their connections with more orthodox mechanistic hypothesis-driven science, as involving three steps:
1. The generation of a preliminary set of hypotheses from an established set of premises; 2. The prioritization of some hypotheses and discarding of others by means of other premises and new evidence; 3. The search for more stringent evidence for prioritized hypotheses. (Ratti 2015 , p. 201)
In step (1), scientific hypothesising plays a role, insofar as it is used to delimit the domain of inquiry of the study. For example, a loss-of-function screen to identify the receptor for a pathogen would hypothesise that there exists a non-redundant mechanism for the interaction of the pathogen with the cells, and that the loss of this cellular factor/mechanism would lead to diminution of interaction of the pathogen with the cell surface. For the purpose of the test, such hypotheses are regarded as indubitable: they delimit the range of acceptable empirical enquiry. But there is also a forward-looking dependence of these approaches on scientific hypothesising: the results of such studies can be used to generate more specific mechanistic hypotheses, certain of which are prioritised in step (2) (based on certain additional assumptions—e.g., that there is a single cellular factor/mechanism responsible for pathogen-cell interaction in the above example), and which can then be validated in downstream analysis in step (3). For example, identification of candidate viral receptors using genome-wide loss-of-function screens can be used to generate specific hypotheses regarding the identity of the associated receptor, which can then be subject to empirical test.
Although broadly speaking we concur with Ratti on these matters (in addition to concurring with other philosophers who have written on this topic, e.g. Franklin ( 2005 ); Burian ( 2007 )), and find his work to deliver significant advances in our conceptual understanding of such large-scale studies, his citing of ‘genome-wide association studies’ (GWAS) as a means of illustrating the above points (see Ratti 2015 , p. 201) invites further consideration. GWAS aims to identify causal associations between genetic variations and diseases/traits; however, it encounters serious difficulties in identifying concrete hypotheses to prioritise, as per Ratti’s (2). Different solutions to this issue (and the related issue of GWAS ‘missing heritability’) manifest in different approaches to this prioritisation: something which deserves to be made explicit in the context of Ratti’s framework. Specifically, while Ratti focuses implicitly on a ‘core gene’ approach to GWAS (cf. Boyle et al. ( 2017 )), according to which a small number of ‘single nucleotide polymorphisms’ (this terminology will be explained in the body of this paper) are primarily responsible for the trait in question (note that this does not imply that only a small number of genes are associated with the relevant phenotype—rather, it assumes that there are some genes which are more central for the manifestation of the phenotype than the majority), there are other approaches to GWAS which do not presuppose this core gene model; as explained in Wray et al. ( 2018 ) (albeit without direct reference to Ratti’s work), such approaches would lead to the prioritisation of different hypotheses in Ratti’s (2). Footnote 1
The first goal of the present paper is to expand on these matters in full detail, and to revise Ratti’s framework in order to incorporate the above points: in so doing, we gain a clearer understanding of how GWAS approaches relate to more traditional, mechanistic, hypothesis-driven science. But there is also a second goal of this paper: to explore for the first time (to our knowledge) in the philosophical literature what it would take for the above-mentioned alternative approaches (often relying on network models)—particularly those which appeal to the field of (phospho)proteomics—to succeed. Although we make no claim that such (phospho)proteomics approaches are per se superior to other strategies for hypothesis prioritisation, they are nevertheless in our view worthy of philosophical attention unto themselves, for they constitute (we contend) a novel form of exploratory experimentation (cf. Burian ( 2007 ); Franklin ( 2005 ); Steinle ( 1997 )) featuring both iterativity (cf. Elliott ( 2012 ); O’Malley et al. ( 2010 )) and appeal to deep learning (cf. Bechtel ( 2019 ); Ratti ( 2020 )).
Bringing all this together, the plan for the paper is as follows. In Sect. " GWAS studies and prioritisation ", we recall the details of GWAS, and witness how different approaches to the so-called missing heritability and coherence problems lead to the prioritisation of different hypotheses in Ratti’s (2). In Sect. " Proteomics and iterative methodology ", we turn our attention to network approaches—specifically to those informed by (phospho)proteomics—and study these through the lens of the literature on exploratory experimentation, before returning to our considerations of GWAS and addressing the question of how such network-based approaches inform the question of hypothesis prioritisation in that context. We close with some discussion of future work to be done in the philosophy both of GWAS, and of big-data biology at large.
GWAS studies and prioritisation
Background on gwas.
Many applications of the framework presented in the introduction—perform genome-wide screens based on a general hypothesis (for example, ‘a gene/process is responsible for a disease’), and on the basis of the results obtained construct a more refined hypothesis for further testing—have been highly successful in biomedical research. However, there are cases in which the application of the approach has not been so straightforward. This can best be illustrated using the example of a field of genomics that studies common diseases such as inflammatory bowel disease (IBD), coronary artery disease, insomnia, and depression. These are often diseases complex in nature, and are thought to be controlled not by a single mutation, but rather to be influenced by multiple loci in the genome and even through the effect of the environment.
In the past decades, researchers have developed a method to characterise the genotype-phenotype associations in these diseases: the method is called ‘genome-wide association studies’ (GWAS). To understand this method, it is important to understand single nucleotide polymorphisms (SNPs). SNPs are variations in a single DNA building block, called a ‘nucleotide’, and they constitute the most common type of genetic variation among individuals. There are around 4-5 million SNPs in a person’s genome. Most SNPs have no effect on human health, but there are some cases in which these variations lead to increased chances of disease. GWAS was based originally upon a ‘common disease, common variant’ hypothesis, which states that common diseases can be attributed to common genetic variants (present in more than 1–5% of the population). By scanning the genomes of many different people, GWAS sought to identify the relationships between common genetic variations and common traits. GWAS studies remain very popular in the field of human genetics, and have been successful in identifying a number of novel variant-trait associations (for example, in diseases such as those mentioned above). For a clear introduction to GWAS from the biology literature, see Tam et al. ( 2019 ); for existing philosophical works on GWAS, with further details on such studies complimentary to those presented in this paper, see e.g. Bourrat ( 2020 ); Bourrat and Lu ( 2017 ).
GWAS’ discontents
GWAS is, however, not without its critics. A clear conclusion from multiple GWAS studies is that even statistically highly significant hits identified from such studies are able to account only for a small fraction of the heritability of the trait/disease in question. (Recall that ‘heritability’ is the measure of proportion of the phenotypic variance in a population that can be attributed to genetic differences—see Downes and Matthews ( 2020 ) and references therein for further details.) Moreover, GWAS studies often implicate large numbers of genes. To put this into perspective, three GWAS studies performed for height in 2008 identified 27, 12 and 20 associated genomic regions, which accounted merely for 3.7, 2.0, and 2.9% of the population variation in height, respectively ( Lettre et al. ( 2008 ); Weedon et al. ( 2008 ); Gudbjartsson et al. ( 2008 )). This was in sharp contrast with estimates from previous genetic epidemiology studies, based upon twin studies, Footnote 2 that estimated the heritability of height to be around 80% (Yang et al. ( 2010 )). In the early days of GWAS, this apparent discrepancy from GWAS came to be known as the missing heritability problem . For recent philosophical discussion of this problem, see Bourrat ( 2020 ); Bourrat and Lu ( 2017 ); Bourrat et al. ( 2017 ); Bourrat ( 2019 ); Downes and Matthews ( 2020 ); Matthews and Turkheimer ( 2019 ).
Geneticists have since proposed a number of solutions to the missing heritibility problem. The three most commonly-discussed such solutions are classified by Gibson ( 2012 ) as follows:
Complex dieseases are polygenic and many loci with small effects account for the phenotype variance.
Common diseases are caused by rare genetic variants each of which have large effect sizes.
Most common diseases are a result of interactions between many factors such as gene-gene interaction effects and effects from environmental factors.
(We take the proposals for solving the missing heritability problem presented in Bourrat ( 2020 ); Bourrat and Lu ( 2017 ); Bourrat et al. ( 2017 ); Bourrat ( 2019 ), which invoke factors from the epigenome, to fall into category (3); we discuss further these proposals in Sect. GWAS reprise .) From multiple GWAS studies on common diseases there is now overwhelming evidence that common diseases are polygenic, as large numbers of genes are often implicated for a given disease. However, using this framework, it is estimated that it would take 90,000–100,000 SNPs to explain 80% of the population variation in height. In light of this, Goldstein ( 2009 ) raised the concern with GWAS studies that “[i]n pointing at ‘everything’, the danger is that GWAS could point at ‘nothing”’.
It is understandable that one would find unpalatable its not being the case that a single gene or process can be associated with a particular disease. But the situation here is not as straightforward as the above remarks might suggest. Indeed, Boyle et al. ( 2017 ) propose the following refinement of this idea:
Intuitively, one might expect disease-causing variants to cluster into key pathways that drive disease etiology. But for complex traits, association signals tend to be spread across most of the genome—including near many genes without an obvious connection to disease. We propose that gene regulatory networks are sufficiently interconnected such that all genes expressed in disease-relevant cells are liable to affect the functions of core disease-related genes and that most heritability can be explained by effects on genes outside core pathways. We refer to this hypothesis as an ‘omnigenic’ model.
Boyle et al. ( 2017 ) propose that within the large number of implicated genes in GWAS, there are a few ‘core’ genes that play a direct role in disease biology; the large number of other genes identified are ‘peripheral’ and have no direct relevance to the specific disease but play a role in general regulatory cellular networks. By introducing their ‘omnigenic’ model, Boyle et al. ( 2017 ) acknowledge the empirical evidence that GWAS on complex diseases does in fact implicate large number of genes; they thereby seem to draw a distinction between complex diseases and classical Mendelian disorders, in which small number of highly deleterious variants drive the disease. However, their suggestion of the existence of a small number of ‘core’ genes backtracks on this and paints complex diseases in the same brushstrokes as classical Mendelian disorders. A number of authors have welcomed the suggestion that genes implicated for complex diseases play a role in regulatory networks but have found the dicotomy between core and peripheral genes to be an ill-motivated attempt to fit complex disease into what we intuitively think should be the framework of a disease (‘a small number of genes should be responsible for a given disease’). For example, Wray et al. ( 2018 ) write:
It seems to us to be a strong assumption that only a few genes have a core role in a common disease. Given the extent of biological robustness, we cannot exclude an etiology of many core genes, which in turn may become indistinguishable from a model of no core genes.
We concur with this verdict. One possible reconstruction of the reasons underlying the endorsement by Boyle et al. ( 2017 ) of ‘core’ versus ’peripheral’ genes could be in order to solve the missing heritability problem. These authors advocate for using experimental methods that are able to identify rare variants that have high effect sizes (solution (2) of the missing heritability problem as presented above), as this is where they suspect the ‘core’ genes can be identified. However, there is at present no evidence that the ‘core gene’ hypothesis need invariably be true for complex diseases (cf. Wray et al. ( 2018 )), so one might be inclined to reject the original hypothesis that all diseases must fit the mould of ‘small number of genes cause complex diseases’. In so doing, one would thereby need to embrace the claim that at least some complex diseases are polygenic and that putative ‘core’ genes are, in fact, no more important than putative ‘peripheral’ genes in this context.
This, however, still leaves us with the original issue that Boyle et al. ( 2017 ) were trying to address: how is it that genes which look disconnected are, in fact, together implicated in a given disease? In addressing this question, we again concur with Wray et al. ( 2018 ), who write:
To assume that a limited number of core genes are key to our understanding of common disease may underestimate the true biological complexity, which is better represented by systems genetics and network approaches.
That is to say, understanding gene functions and the interplay between the different genes is key to answering why many genes are involved in complex diseases. This is not a straightforward task and a full characterisation of the roles that genes play in biological systems remains a distant prospect.
One approach to addressing this issue is to identify relationships between genes in a cell by way of a systems biology approach, underlying premises of which are that cells are complex systems and that genetic units in cells rarely operate in isolation. Hence, on this view, understanding how genes relate to one another in a given context is key to establishing the true role of variants identified from GWAS hits. There are a number of approaches described in the field of systems biology to identify gene-gene relationships. One widely-implemented approach is to construct ‘regulatory networks’ relating these genes. A regulatory network is a set of genes, or parts of genes, that interact with each other to control a specific cell function. With recent advances in high-throughput transcriptomics, it is now possible to generate complex regulatory networks of how genes interact with each other in biological processes and define the roles of genes in a context-dependent manner based on mRNA expression in a cell. As the majority of GWAS hits often lie in non-coding regions of the genome, which are often involved in regulating gene expressions, networks based on mRNA expression are powerful means to interpret of the functional role of variants identified by GWAS.
Another approach to the functional validation of GWAS hits—currently substantially less common—proceeds by constructing networks generated from expression of proteins/phosphoproteins in a cell (more details of these approaches will be provided in the following section). Such approaches would in principle depict completely the underlying state of the cell. Combined with gene expression data, protein expression networks and signalling networks from proteomics would make transparent the functional role of the variants identified in GWAS studies in a given context—that is, they would provide a mechanistic account of disease pathogenesis without recourse to a neo-Mendelian ‘core gene’ model. Genes which prima facie appear disconnected and irrelevant to disease biology may be revealed by these approaches to be relevant after all. To illustrate, consider a complex disease such as IBD: it is thought that both (i) a disturbed interaction between the gut and the intestinal microbiota, and (ii) an over-reaction of the immune system, are required for this disease phenotype to manifest. Thus, it is likely that a number of genetic pathways will be important—pathways which need not prima facie be connected, but which may ultimately be discovered to be related in some deeper way. These proteomics-informed network approaches would thereby afford one resolution to what has been dubbed by Reimers et al. ( 2019 ) and Craver et al. ( 2020 ) the ‘coherence problem’ of GWAS: to explain how it is that all genes implicated in these studies are related to one another mechanistically. Footnote 3 Clearly, these approaches could be brought to bear in order to vindicate responses (1) or (3) to the missing heritability problem, presented above. Footnote 4
To close this subsection, it is worth reflecting on how the ‘core gene’ hypothesis might intersect with network-based approaches. If a core gene exists, then a network analysis should (at least in principle) be able to identify it; in this sense, a ‘core gene’ hypothesis can be compatible with a network approach. As already mentioned above, however, there is no evidence that such core genes invariably exist: a network analysis could (in principle) identify many ‘central hubs’, rather than just one—an outcome not obviously compatible with the ‘core gene’ hypothesis. (For more on this latter possibility, cf. the very recent work of Barrio-Hernandez et al. ( 2021 ), discussed further below.)
Ratti’s framework for large-scale studies
Suppose that one follows (our reconstruction of) Boyle et al. ( 2017 ), in embracing option (2) presented above as a solution to the GWAS missing heritability problem. One will thereby, in Ratti’s second step in his three-step programme characterising these data-driven approaches to biology, prioritise hypotheses according to which a few rare genes are responsible for the disease in question. This, indeed, is what Ratti ( 2015 ) suggests in §2.2 of his article. However, one might question whether this prioritisation is warranted, in light of the lack of direct empirical evidence for this neo-Mendelian hypothesis (as already discussed). Wray et al. ( 2018 ), for example, write that
... [t]o bias experimental design towards a hypothesis based upon a critical assumption that only a few genes play key roles in complex disease would be putting all eggs in one basket.
If one concurs with Wray et al. ( 2018 ) on this matter (as, indeed, we do), then one may prioritise different hypotheses in the second step of Ratti’s programme—in particular, one may prioritise specific hypotheses associated with ‘polygenic’ models which would constitute approach (1) and/or approach (3) to the missing heritability problem.
This latter point should be expanded. Even if one does embrace a ‘polygenic’ approach to the missing heritability problem (i.e., approach (1) and/or approach (3)), and applies e.g. networks (whether transcriptomics-based, or (phospho)proteomics-informed, or otherwise—nothing hinges on this for our purposes here) in order to model the genetic factors responsible for disease pathogenesis, ultimately one must prioritise specific hypotheses for laboratory test. For example, Schwartzentruber et al. ( 2021 ) implement in parallel a range of network models within the framework of a polygenic approach in order to prioritise genes such as TSPAN14 and ADAM10 in studies on Alzheimer’s disease (we discuss further the methodology of Schwartzentruber et al. ( 2021 ) in §3.3 ). Note, however, that these specific hypotheses might be selected for a range of reasons—e.g., our prior knowledge of the entities involved, or ease of testability, or even financial considerations—and that making such prioritisations emphatically does not imply that one is making implicit appeal to a ‘core gene’ model. This point is corroborated further by the fact that the above two genes are not the most statistically significant hits in the studies undertaken by Schwartzentruber et al. ( 2021 ), as one might expect from those working within the ‘core gene’ framework.
Returning to Ratti’s framework: we take our noting this plurality of options vis-à-vis hypothesis prioritisation to constitute a friendly modification to this framework appropriate to contexts such as that of GWAS. But of course, if one were to leave things here, questions would remain—for it would remain unclear which polygenic model of disease pathogenesis is to be preferred, and how such models are generated. Given this, it is now incumbent upon us to consider in more detail how such approaches set about achieving these tasks in practice: due both to their potential to offer underlying mechanistic models of the cell, as well as due to the novel iterative methodology for hypothesis generation involved, we focus largely in the remainder upon (phospho)proteomics-based approaches.
Proteomics and iterative methodology
Proteomics promises to afford the ultimate fundamental mechanistic account of cellular processes; data from proteomics would, therefore, illuminate the underlying relationships between the variants identified in GWAS studies. In this section, we explore in greater detail how such proteomics approaches proceed; they constitute a novel form of ‘exploratory experimentation’ (in the terminology of Burian ( 2007 ); Steinle ( 1997 )) worthy unto themselves of exposure in the philosophical literature. Footnote 5 In proteomics, further complications for hypothesis generation and testing arise, for data is sparse, and experiments often prohibitively expensive to perform. Given these constraints, how is progress to be made? It is to this question which we now turn; the structure of the section is as follows. In Sect. Proteomics: a data-deprived field , we present relevant background regarding proteomics. Then, in Sect. Methodological iteration , we argue that the development of this field can be understood on a model of a novel form of iterative methodology (cf. Chang 2004 ; O’Malley et al. 2010 ). We return to the relevance of these approaches for GWAS in Sect. GWAS reprise .
Proteomics: a data-deprived field
The ultimate aim of -omics studies is to understand the cell qua biological system. Transcriptomics is now sufficiently well-advanced to accommodate large-scale systematic studies to the point of being used to validate variants identified from GWAS. Footnote 6 By contrast, proteomics—the study of proteins in a cell—remains significantly under-studied. Technologies allowing for the systematic study of proteins are not as advanced as those for studying genes and transcripts; this is mainly because no method currently exists for directly amplifying proteins (i.e., increasing the amount of a desired protein in a controlled laboratory context): a methodology which has been key for genomics and transcriptomics. Proteins are very diverse in the cell: a single gene/transcript gives rise to multiple proteins. Proteins themselves can be modified in the cell after being created, thus further increasing the complexity of proteomics studies. Unlike genomics and transcriptomics, in which it is now common to perform systematic genome-wide or transcriptome-wide approaches, studies of proteins are therefore usually taken piecemeal.
Proteomics research tends to focus on families of proteins that are involved in a particular known biological process. Among the important families of proteins are kinases and phosphatases, which are molecules that are responsible for signal transmission in the cell. These proteins are able to modify other proteins by adding or removing a phosphate group (respectively). This modification changes the shape (‘conformation’) of the protein, rendering it active or inactive, Footnote 7 depending on the context. By examining the phopsphorylation state of the proteins inside a cell, it is possible to infer the signalling state of that cell. The field of phosphoproteomics aims to characterise all phospho-modified proteins within a cell. This is thought to be one of the most powerful and fundamental ways of inferring the signalling process within a cell; the approach could add a substantial new layer to our understanding of both basic and disease biology. That said, a recent estimate suggests that current approaches have identified kinases for less than 5% of the phosphoproteome. What is even more staggering is that almost 90% of the phosphorylation modifications that have been identified have been attributed to only 20% of kinases. The other 80% of the kinases are completely dark: their functions remain unknown. For many such kinases, we do not even know where in the cell they are located. (See Needham et al. ( 2019 ) for a review of the current state of play in phosphoproteomics.)
In such a field, systematic studies to quantify the entire phosphoproteome in a cell and an ability to assign a kinase to every phosphorylated component would be the ultimate aim. But phosphoproteomics studies themselves are currently extremely expensive, and there are technological limitations in mapping the global phosphoproteome—not least sparsity of data, which often comes as a result of limitations in the technical setup of laboratory measurements and experiments. For example: the same sample measured in the same machine at two different instances will give readings for different phosphoproteins. Some statistical methods can be used to overcome these limitations, but these require making assumptions regarding the underlying biology, which defeats the point of an unbiased study.
In spite of these difficulties, it has been shown that if one combines multiple large-scale phosphoprotemics data sets (each admittedly incomplete), it is possible to predict kinase-kinase regulatory relationships in a cell using data-driven phosphoprotein signalling networks obtained via supervised machine learning approaches (a recent study from Invergo et al. 2020 showcases one such approach; we will use this as a running example in the ensuing). Footnote 8 First, a training set of data is used to teach a machine a classification algorithm. Once the classification algorithm is learnt, the machine is set to the task of applying it to unlabelled data: in our case, the goal is to identify further, as-yet unknown, regulatory protein relationships or non-relationships. (On machine learning and network analysis of biological systems, see also Bechtel ( 2019 ) and Ratti ( 2020 ).)
Before assessing such phosphoproteomics machine learning algorithms as that of Invergo et al. ( 2020 ), there are two further complications with the current state of play in proteomics which need to be mentioned. First: it is much easier to curate positive lists of interactions than negative lists. (This is essentially a case of its being easier to confirm existentially quantified statements than universally quantifies statements: for how can we ever truly ascertain that any two given proteins never interact?) Thus, at present, negative lists obtained from laboratory experiments are underpopulated. Invergo et al. ( 2020 ) attempt to circumvent this issue in the following way: they assume that regulatory relationships are rare, so that if one were to randomly sample protein associations, one could create reliably large artificial negative sets; indeed, they do generate artificial negative sets in exactly this way. (Clearly, this means that these approaches again cannot be understood as being ‘hypothesis-free’: cf. Sect. Introduction .)
The second problem with the current state of play in proteomics is this: when a given interaction occurs is a function of multifarious factors, most notably cell context. This context-dependence means that an entry in a negative set in one context might, in fact, be an entry in a positive set in another. To illustrate: in the case of regulatory relationships between two kinases, it is known that such relationships can be prone to dysregulation in diseases such as cancer. Hence, a well-annotated positive set relationship can very well be dysregulated in a cancer context, so that this relationship no longer exists, effectively putting it into a negative set. The problem is that many data-driven approaches rely on data that are generated in simple reductionist systems such as cancer cell lines—so that the results obtained might not carry across to the target physiological context. (Cancer cell lines can grow infinitely, and thus are ideal for experiments.) The approach taken by Invergo et al. ( 2020 ) utilises data from breast cancer cell lines; hence, the relationships they predict could be specific to a dysregulated system. In response to this second problem, we suggest replying on behalf of Invergo et al. ( 2020 ) that most regulatory relationships fundamental to the functioning of the cell should hold true in most contexts. At present, however, given the data-deprived nature of proteomics, there is little direct evidence for this hypothesis. (Again, the appeal to any such hypothesis would mean that such proteomics approaches cannot be ‘hypothesis-free’.)
Thus, the fact that Invergo et al. ( 2020 ) utilise data from breast cancer cell lines raises the possibility that their machine learning algorithms might be trained on data unsuited to other contexts, leading to concerns regarding error propagation. This general concern regarding the context-specificity (or lack thereof) of input data sets is, however, recognised by authors in the field—for example, Barrio-Hernandez et al. ( 2021 ) note that “improvements in mapping coverage and computational or experimental approaches to derive tissue or cell type specific networks could have a large impact on future effectiveness of network expansion” (Barrio-Hernandez et al. 2021 , p. 14).
Methodological iteration
In spite of these problems, Invergo et al. ( 2020 ) argue that the results obtained from their approach afford a useful means of bootstrapping further progress in phosphoproteomics. As they put it:
Although we do not suggest that these predictions can replace established methods for confirming regulatory relationships, they can nevertheless be used to reduce the vast space of possible relationships under consideration in order to form credible hypotheses and to prioritize experiments, particularly for understudied kinases. (Invergo et al. 2020 , p. 393)
One way to take this point is the following. Ideally, in order to construct positive and negative sets, one would test in the laboratory each individual protein association. Practically, however, this would be an unrealistic undertaking, as we have already seen. What can be done instead is this:
Generate a global phosphoproteomics data set, albeit one that is incomplete and sparse (e.g., that presented in Wilkes et al. ( 2015 )), based upon laboratory experiments.
Train, using this data set and input background hypotheses of the kind discussed above, a machine learning algorithm (such as that presented in Invergo et al. ( 2020 )) to identify candidate interactions in the unknown space of protein-protein interactions. Footnote 9
Use these results to guide further laboratory experimentation, leading to the development of more complete data sets.
Train one’s machine learning algorithms on these new data sets, to improve performance; in turn, repeat further the above process.
Clearly, a process of reflective equilibrium is at play here (cf. Daniels ( 2016 )). As is well-known, Chang ( 2004 ) has proposed an iterative conception of scientific methodology, according to which the accrual of scientific hypotheses is not a linear matter; rather, initial data may lead to the construction of a theoretical edifice which leads one to develop new experiments to revise one’s data; at which point, the process iterates. This fits well with the above-described procedures deployed in phosphoproteomics; it also accords with previous registration of the role of iterative procedures in large-scale biological studies—see e.g. O’Malley et al. ( 2010 ) and Elliott ( 2012 ).
Let us delve into this a little deeper. As Chang notes,
There are two modes of progress enabled by iteration: enrichment , in which the initially affirmed system is not negated but refined, resulting in the enhancement of some of its epistemic virtues; and self-correction , in which the initially affirmed system is actually altered in its content as a result of inquiry based on itself. (Chang 2004 , p. 228)
Certainly and uncontroversially, enrichment occurs in the above four-step process in phosophoproteomics: the new data yield a refinement of our previous hypotheses in the field. In addition, however, it is plausible to understand the above iterative methodology as involving self-correction: for example, in might be that the machine learning algorithm of Invergo et al. ( 2020 ) identifies a false positive, yet nevertheless makes sufficiently focused novel predictions with respect to other candidate interactions in order to drive new experimentation, leading to a new data set on which the algorithm can be trained, such that, ultimately, the refined algorithm does not make a false positive prediction for that particular interaction. This is entirely possible in the above iterative programme; thus, we maintain that both modes of Changian iterative methodology are at play in this approach.
There is another distinction which is also relevant here: that drawn by Elliott ( 2012 ) between ‘epistemic iteration’—“a process by which scientific knowledge claims are progressively altered and refined via self-correction or enrichment”—and ‘methodological iteration’—“a process by which scientists move repetitively back and forth between different modes of research practice” (Elliott 2012 , p. 378). It should be transparent from our above discussion that epistemic iteration is involved in these proteomics approaches. Equally, though, it should be clear that methodological iteration is involved, for the approach alternates between machine learning and more traditional laboratory experimentation. That machine learning can play a role in an iterative methodology does not seem to have been noted previously in the philosophical literature—for example, it is not identified by Elliott ( 2012 ) as a potential element of a methodologically iterative approach; on the other hand, although the role of machine learning in network modelling and large-scale studies is acknowledged by Bechtel ( 2019 ) and Ratti ( 2020 ) (the latter of whom also discusses—albeit without explicitly using this terminology—the role of machine learning in epistemic iteration: see (Ratti 2020 , p. 89)), there is no mention of its role in an iterative methodology such as that described above.
GWAS reprise
Given the foregoing, we hope it is reasonable to state that the approaches to proteomics of e.g. Invergo et al. ( 2020 ) constitute novels forms of exploratory experimentation, worthy of study in their own right. Let us, however, return now to the matter of polygenic approaches to GWAS hits. In principle, the results of the methodologies of e.g. Invergo et al. ( 2020 ) could further vindicate these approaches, by providing mechanistic models of which genes interact in a disease context, and when and why they do so. In turn, they have the capacity to allow biologists to prioritise specific hypotheses in Ratti’s step (2), without falling back upon assumptions that only few genes are directly involved in complex disease biology.
Note that that there is a complex interplay between this iterative methodology and the ‘eliminative induction’ of stages (1) and (2) Ratti’s analysis (see Sect. Introduction ; for earlier sources on eliminative induction, see Earman ( 1992 ); Kitcher ( 1993 ); Norton ( 1995 )). We take this to consist in the following. First, a methodology such as that of Invergo et al. ( 2020 ) is used to generate a particular network-based model for the factors which are taken to underlie a particular phenotype. This model is used to prioritise ( à la eliminative induction) particular hypotheses, as per stage (2) of Ratti’s framework; these are then subject to specific test, as per stage (3) of Ratti’s framework. The data obtained from such more traditional experimentation is then used to construct more sophisticated network models within the framework of Invergo et al. ( 2020 ); these in turn lead to the (eliminative inductive) prioritisation of further specific hypotheses amenable to specific test. As already discussed above, this is a clear example of the ‘methodological iteration’ of Elliott ( 2012 ).
It bears stressing that (phospho)proteomics network-based approaches may, ultimately, constitute only one piece of the solution to the broader puzzle that is GWAS hypothesis prioritisation. In very recent work, Schwartzentruber et al. ( 2021 ) have brought to bear upon this problem consideration of, inter alia , epigenomic factors alongside network-based analyses. There are two salient points to be made on this work. First: although Bourrat et al. ( 2017 ) are correct that epigenomic studies and background may have a role to play in addressing the missing heritability problem (cf. Bourrat ( 2019 , 2020 ); Bourrat and Lu ( 2017 )), a view in contemporary large-scale biological studies—evident in papers such as Schwartzentruber et al. ( 2021 )—is that these considerations can be supplemented with yet other resources, such as network-based studies; we concur with this verdict. Second: in order to construct these networks, Schwartzentruber et al. ( 2021 ) rely on established protein-protein interaction databases such as STRING, IntAct and BioGRID (Schwartzentruber et al. 2021 , p. 397). While effective in their own right, networks developed from such databases have the disadvantage that they represent signalling in an ‘average’ cell, and are therefore unsuitable for studying dynamic context- and cell-type-specific signalling responses (cf. Sharma and Petsalaki ( 2019 )). In this regard, it would (at least in principle) be preferable to utilise regulatory and context-specific networks developed using methods described in work such as that of Invergo et al. ( 2020 ) in future approaches to GWAS hypothesis prioritisation. That being said, in practice this may not yet be fruitful, as at present contemporary large-scale biology is only at the early stages of the iterative processes discussed above; moreover, the training data sets used by such methods remain at this stage not completely context-specific (recall that Invergo et al. ( 2020 ) utilise a breast cancer training set)—meaning that the potential of such work to yield detailed, context-specific network-based models is yet to be realised in full.
With all of the above in hand, we close this subsection by considering more precisely the question of how the machine learning algorithms of Invergo et al. ( 2020 ) bear upon the missing heritability problem. Having developed regulatory protein-protein interaction networks on the basis of such algorithms, one can take (following here for the sake of concreteness the lead of Barrio-Hernandez et al. ( 2021 )) the connection with hypothesis prioritisation in GWAS (and, in turn, the missing heritability problem) to proceed via the following steps (also summarised visually in Fig. 1 ):
Select a protein-protein interaction network. Usually, this is a pre-existing curated network, such as those defined in the STRING database (discussed above). However, instead of such curated networks, use in their place networks developed on the machine learning models of e.g. Invergo et al. ( 2020 ).
Within those networks, identify the nodes (i.e., proteins) which correspond to hits from a particular GWAS (i.e., the proteins associated with the genes identified in the GWAS). Footnote 10
Use network propagation methods (see e.g. Cowen et al. ( 2017 ) for a review of such methods), potentially alongside other factors (as discussed in e.g. Schwartzentruber et al. ( 2021 )) in order to identify known modules (i.e., separated substructures within a network) associated with the disease in question.
Target elements of those modules, regardless of whether or not they were hits in the original GWAS. (This latter approach—of targeting beyond the original GWAS hits—is novel to the very recent work of Barrio-Hernandez et al. ( 2021 ).)
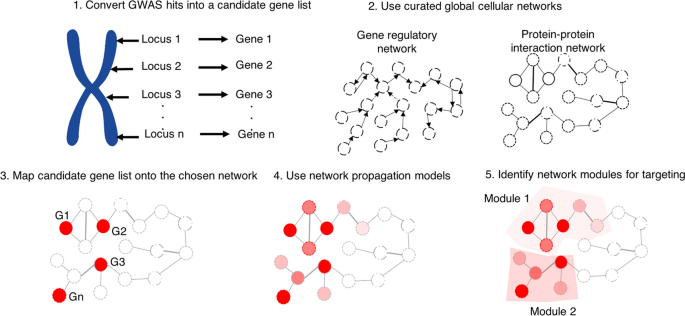
The application of networks to GWAS hit prioritisation. In (1), GWAS hits are converted to candidate gene lists. In (2), one selects a cellular network: this could be a gene regulatory network, or a protein-protein interaction network (e.g. from STRING), or a protein-protein regulatory network (possibly constructed via the machine learning methodologies of Invergo et al. ( 2020 )). In (3), genes associated with the GWAS loci are mapped to the chosen network. In (4), network propagation methods (e.g. diffusion techniques) are applied in order identify potential disease-related genes not picked up by the GWAS. In (5), the results of these network analyses are used to identify significant genetic modules to be targeted experimentally in investigations into disease pathogenesis. Note, following Wray et al. ( 2018 ) and Barrio-Hernandez et al. ( 2021 ), that this particular means of bridging the gap between cellular networks and investigations into the results of GWAS hits does not presuppose a ‘core gene’ hypothesis
On (2) and (3): Boyle et al. ( 2017 ) may or may not be correct that many genes are implicated (either in the original screen, or after the network analysis has been undertaken)—recall from Sect. GWAS’ discontents their ‘omnigenic’ model. However, on the basis of the work of Barrio-Hernandez et al. ( 2021 ) one might argue that this is not the most important question—rather, the important question is this: which gene modules provide insights into the disease mechanism? One can ask this question without subscribing to a ‘core gene’ model; thus, we take the work of Barrio-Hernandez et al. ( 2021 ) to be consistent with the above-discussed points raised by Wray et al. ( 2018 ).
This paper has had two goals. The first has been to propose revisions to the framework of Ratti ( 2015 ) for the study of the role of hypothesis-driven research in large-scale contemporary biological studies, in light of studies such as GWAS and its associated missing heritability problem. In this regard, we have seen that different hypotheses may be prioritised, depending upon whether one adopts a ‘core’ gene model (as Ratti ( 2015 ) assumes, and as is also advocated in Boyle et al. ( 2017 )), or whether one adopts a polygenic model (as endorsed by Wray et al. ( 2018 ); cf. Barrio-Hernandez et al. ( 2021 )). The second goal of this paper has been to consider how these hypotheses would be developed on polygenic approaches via (phospho)proteomics—which itself constitutes a novel form of exploratory experiment, featuring as it does both iterativity and deep learning—and to consider what it would take for these network-based proteomics approaches to succeed. A broader upshot of this paper has been the exposure for the first time to the philosophical literature of proteomics: given its potential to provide mechanistic models associated with disease phenotypes, the significance of this field cannot be overstated.
The issues discussed in this paper raise important questions regarding how researchers prioritise not just first-order hypotheses as per Ratti’s (2), but also the background assumptions which allow one to make such adjudications to begin with. To be concrete: in the case of GWAS, should one prioritise the assumption that rare variants of large effect in a small number of genes drive complex diseases, or rather invest in developing systems-based approaches and in improving under-studied fields, such as (phospho)proteomics, which may or may not ultimately shed light on the question of why complex diseases have thus far manifested empirically as polygenic? These choices lead to different first-order prioritisations in Ratti’s second step, and thereby have great potential to steer the course of large-scale studies in future years. Given limited resources in the field, it is, in our view, worth pausing to reflect on whether said resources are appropriately allocated between these options, and to strive to avoid any status quo bias in favour of currently-popular assumptions. Footnote 11
In fairness to Ratti, in other articles, e.g. López-Rubio and Ratti ( 2021 ), he does not make assumptions tantamount to a ‘core gene’ hypothesis; in this sense, our criticism falls most squarely on assumptions made in Ratti ( 2015 ).
Twin studies are powerful approaches to studying the genetics of complex traits. In simple terms, twin studies compare the phenotypic similarity of identical (monozygotic) twins to non-identical (dizygotic) twins. As monozygotic twins are genetically identical and non-identical twins are on average ‘half identical’, observing greater similarity of identical over non-identical twins can be used as an evidence to estimate the contribution of genetic variation to trait manifestation. For further discussion of twin studies in the philosophical literature, see e.g. Matthews and Turkheimer ( 2019 ); Downes and Matthews ( 2020 ).
There are many further questions to be addressed here in connection with the literature of mechanisms and mechanistic explanations. For example, are these network approaches best understood as revealing specific mechanisms, or rather as revealing mechanism schema (to use the terminology of (Craver and Darden 2013 , ch.3))? Although interesting and worthy of pursuit, for simplicity we set such questions aside in this paper, and simply speak of certain contemporary biology approaches as revealing ‘underlying mechanisms’. In this regard, we follow the lead of Ratti ( 2015 ).
To be completely clear: we do not claim that these (phospho)proteomics-based network approaches are superior to regulatory network approaches, given the current state of technology in the field. On the contrary—as we explain in Sect. Proteomics and iterative methodology —the former of these fields is very much nascent, and has yet to yield significant predictive or explanatory fruit. Nevertheless—again as we explain in Srct. Proteomics and iterative methodology —in our view these approaches are worthy of exposure in the philosophical literature in their own right, for (a) they offer one of the most promising means (in principle, if not yet in practice) of providing a mechanistic account of disease pathogenesis, and (b) the particular way in which hypotheses are developed and prioritised on these approaches is conceptually rich.
Recall: “Experiments count as exploratory when the concepts or categories in terms of which results should be understood are not obvious, the experimental methods and instruments for answering the questions are uncertain, or it is necessary first to establish relevant factual correlations in order to characterize the phenomena of a domain and the regularities that require (perhaps causal) explanation” (Burian 2013 ). Cf. e.g. Franklin ( 2005 ); Steinle ( 1997 ). All of the -omics approaches discussed in this paper were identified in Burian ( 2007 ) as cases of exploratory experimentation; the details of contemporary proteomics approaches have, however, not been presented in the philosophical literature up to this point (at least to our knowledge).
In this paper, we do not go into the details of specific transcriptomics studies. One interesting approach worthy of mention, however, is ‘single-cell RNA sequencing’ (SC-RNA), which allows biologists to assay the full transcriptome of hundreds of cells in an unbiased manner (see e.g. Hwang et al. ( 2018 ) for a recent review). The advantage of SC-RNA over older methods lies in its ability to identify the transcriptomes from heterocellular and poorly-classified tissue populations and disease-associated cell states.
As the addition or removal of phosphate groups regulates the activity of a protein, such relationships between a kinase and its target (also called a ‘substrate’) are referred to as ‘regulatory relationships’. Kinases themselves can also be phosphorylated by other kinases, so there exist also kinase-kinase regulatory relationships in a cell.
Supervised machine learning involves training a machine on a given data set (for example, a collection of cat photos versus dog photos), before assigning the machine the task of classifying entries in some new data set. By contrast, in unsupervised learning, the machine is instructed to find its own patterns in a given data set. For some recent philosophical considerations regarding machine learning, see Sullivan ( 2019 ).
One can also test the results of the machine binary classification algorithm on other data sets: this Invergo et al. ( 2020 ) did with reference to the data presented in Hijazi et al. ( 2020 ). The design of the algorithmic system and algorithm used by Invergo et al. ( 2020 ) is described with admirable clarity at (Invergo et al. 2020 , pp. e5ff.), to which the reader is referred for further details.
Note that identification of candidate genes from the loci which constitute GWAS hits is non-trivial. The recently-described ‘locus-to-gene’ (L2G) approach is a machine learning tool which can be used to prioritise likely causal genes at each locus given genetic and functional genomics features (see Mountjoy et al. ( 2020 )).
Cf. Samuelson and Zeckhauser ( 1988 ). For related discussion of funding decisions in the context of -omics studies, see Burian ( 2007 ).
Barrio-Hernandez I, Schwartzentruber J, Shrivastava A, del Toro N, Zhang Q, Bradley G, Hermjakob H, Orchard S, Dunham I, Anderson CA, Porras P, Beltrao P (2021) ‘Network expansion of genetic associations defines a pleiotropy map of human cell biology’, bioRxiv . https://www.biorxiv.org/content/early/2021/07/19/2021.07.19.452924
Bechtel W (2019) Hierarchy and levels: analysing networks to study mechanisms in molecular biology. Philos Transact R Soc B 375(20190320):20190320
Google Scholar
Bourrat P (2019) Evolutionary transitions in heritability and individuality. Theory Biosci 138:305–323
Article Google Scholar
Bourrat P (2020) Causation and single nucleotide polymorphism heritability. Philos Sci 87:1073–1083
Bourrat P, Lu Q (2017) Dissolving the Missing Heritability Problem. Philosophy of Science 84:1055–1067
Bourrat P, Lu Q, Jablonka E (2017) Why the missing heritability might not be in the DNA. BioEssays 39:1700067
Boyle E, Li Y, Pritchard J (2017) An expanded view of complex traits: from polygenic to omnigenic. Cell 169:1177–1186
Burian R (2013) Exploratory experimentation. In: Dubitzky W, Wolkenhauer O, Cho K-H, Yokota H (eds) Encyclopedia of systems biology. Springer, Berlin
Burian RM (2007) On MicroRNA and the need for exploratory experimentation in post-genomic molecular biology. Hist Philos Life Sci. 29(3):285–311. http://www.jstor.org/stable/23334263
Chang H (2004) Inventing temperature: measurement and scientific progress. Oxford University Press, Oxford
Book Google Scholar
Cowen L, Ideker T, Raphael BJ, Sharan R (2017) Network propagation: a universal amplifier of genetic associations. Nat Rev Genet 18(9):551–562. https://doi.org/10.1038/nrg.2017.38
Craver CF, Darden L (2013) In search of mechanisms. University of Chicago Press, Chicago
Craver CF, Dozmorov M, Reimers M, Kendler KS (2020) Gloomy prospects and roller coasters: finding coherence in genome-wide association studies. Philos Sci 87(5):1084–1095
Daniels N (2016) Reflective equilibrium. The Stanford Encyclopedia of Philosophy
Doudna JA, Charpentier E (2014) The new frontier of genome engineering with CRISPR-Cas9. Science 346(6213):1258096
Downes SM, Matthews L (2020) Heritability. In: Zalta EN (ed) The Stanford encyclopedia of philosophy. Stanford University, Metaphysics Research Lab
Earman J (1992) Bayes or bust? a critical examination of BayesianConfirmation Theory,. MIT Press, Cambridge
Elliott KC (2012) Epistemic and methodological iteration in scientific research. Stud Hist Philos Sci Part A 43(2):376–382
Franklin L (2005) Exploratory experiments. Philos Sci. 72(5):888–899. https://www.jstor.org/stable/10.1086/508117
Gibson G (2012) Rare and common variants: twenty arguments. Nat Rev Genet 13(2):135–145
Goldstein D (2009) Common genetic variation and human traits. N Engl J Med 360:1696–1698
Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, Sulem P, Thorlacius S, Gylfason A, Steinberg S, Helgadottir A, Ingason A, Steinthorsdottir V, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Pedersen O, Aben KK, Witjes JA, Swinkels DW, Heijer Md, Franke B, Verbeek ALM, Becker DM, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Gulcher J, Kiemeney LA, Kong A, Thorsteinsdottir U, Stefansson K (2008) Many sequence variants affecting diversity of adult human height. Nat Genet 40(5):609–615. https://doi.org/10.1038/ng.122
Hasin Y, Seldin M, Lusis A (2017) Multi-omics approaches to disease. Genome Biol 18(1):83
Hijazi M, Smith R, Rajeeve V, Bessant C, Cutillas PR (2020) Reconstructing kinase network topologies from phosphoproteomics data reveals cancer-associated rewiring. Nat Biotechnol 38(4):493–502
Hwang B, Lee JH, Bang D (2018) Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med 50(8):96. https://doi.org/10.1038/s12276-018-0071-8
Invergo BM, Petursson B, Akhtar N, Bradley D, Giudice G, Hijazi M, Cutillas P, Petsalaki E, Beltrao P (2020) Prediction of signed protein kinase regulatory circuits. Cell Syst 10(5):384-396.e9
Kitcher PS (1993) The advancement of science. Oxford University Press, Oxford
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blöcker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts, P, Koonin, E V, Korf I, Kulp, D, Lancet D, Lowe T M, McLysaght A, Mikkelsen T, Moran J V, Mulder N, Pollara V J, Ponting C P, Schuler G, Schultz J, Slater G, Smit A F, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf Y I, Wolfe, K H, Yang S P, Yeh R F, Collins F, Guyer M S, Peterson J, Felsenfeld A, Wetterstrand K A, Patrinos A, Morgan M J, de Jong P, Catanese J J, Osoegawa K, Shizuya H, Choi S, Chen Y J, Szustakowki J, and International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409(6822):860–921
Leonelli S (2016) Data-centric biology: a philosophical study. University of Chicago Press, Chicago
Lettre G, Jackson A. U., Gieger C., Schumacher F. R., Berndt S. I., Sanna S., Eyheramendy S., Voight B. F., Butler J. L., Guiducci C., Illig T., Hackett R., Heid I. M., Jacobs K. B., Lyssenko V., Uda M., Boehnke M., Chanock S. J., Groop L. C., Hu F. B., Isomaa B., Kraft P., Peltonen L., Salomaa V., Schlessinger D., Hunter D. J., Hayes R. B., Abecasis G. R., Wichmann H.-E., Mohlke K. L., Hirschhorn J. N., Initiative T. D. G., FUSION, KORA, The Prostate, LC, Trial OCS, Study TNH, SardiNIA (2008) Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet 40(5):584–591. https://doi.org/10.1038/ng.125
López-Rubio E, Ratti E (2021) Data science and molecular biology: prediction and mechanistic explanation. Synthese 198(4):3131–3156. https://doi.org/10.1007/s11229-019-02271-0
Matthews LJ, Turkheimer E (2019) Across the great divide: pluralism and the hunt for missing heritability. Synthese. https://doi.org/10.1007/s11229-019-02205-w
Mountjoy E, Schmidt EM, Carmona M, Peat G, Miranda A, Fumis L, Hayhurst J, Buniello A, Schwartzentruber J, Karim MA, Wright D, Hercules A, Papa E, Fauman E, Barrett JC, Todd JA, Ochoa D, Dunham I, Ghoussaini M (2020) Open targets genetics: an open approach to systematically prioritize causal variants and genes at all published human GWAS trait-associated loci, bioRxiv . https://www.biorxiv.org/content/early/2020/09/21/2020.09.16.299271
Needham E, Parker B, Burykin T, James D, Humphreys S (2019) Illuminating the dark phosphoproteome, Sci Signal. Vol. 12
Norton J (1995) Eliminative induction as a method of discovery: how Einstein discovered general relativity. In: Leplin J (ed) The creation of ideas in physics. Kluwer, Alphen aan den Rijn, pp 29–69
Chapter Google Scholar
O‘Malley M, Elliott K, Burian R (2010) From genetic to genomic regulation: iterativity in microRNA research. Stud Hist Philos Sci Part C Stud Hist Philos Biol Biomed Sci 41(4):407–417
Ratti E (2015) Big Data biology: between eliminative inferences and exploratory experiments. Philos Sci 82:198–218
Ratti E (2020) What kind of novelties can machine learning possibly generate? The case of genomics. Stud Hist Philos Sci Part A. 83:86–96. https://www.sciencedirect.com/science/article/pii/S0039368119302924
Reimers M, Craver C, Dozmorov M, Bacanu S-A, Kendler K (2019) The coherence problem: finding meaning in GWAS complexity. Behav Genet 49:187–195
Richardson S, Stevens H (2015) Postgenomics: perspectives on biology after the genome. Duke University Press, Durham
Samuelson W, Zeckhauser R (1988) Status quo bias in decision making. J Risk Uncertain 1(1):7–59. https://doi.org/10.1007/BF00055564
Schwartzentruber J, Cooper S, Liu JZ, Barrio-Hernandez I, Bello E, Kumasaka N, Young AMH, Franklin RJM, Johnson T, Estrada K, Gaffney DJ, Beltrao P, Bassett A (2021) Genome-wide meta-analysis, fine-mapping and integrative prioritization implicate new Alzheimer’s disease risk genes. Nat Genet 53(3):392–402. https://doi.org/10.1038/s41588-020-00776-w
Shalem O, Sanjana NE, Zhang F (2015) High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet 16(5):299–311
Sharma S, Petsalaki E (2019) Large-scale datasets uncovering cell signalling networks in cancer: context matters. Curr Opin Genet Dev. 54:118–124 Cancer Genomics. https://www.sciencedirect.com/science/article/pii/S0959437X18301278
Steinle F (1997) Entering new fields: exploratory uses of experimentation. Philos Sci. 64:S65–S74. http://www.jstor.org/stable/188390
Sullivan E (2019) Understanding from machine learning models. British J Philos Sci
Tam V, Patel N, Turcotte M, Bossé Y, Paré G, Meyre D (2019) Benefits and limitations of genome-wide association studies. Nat Rev Genet 20(8):467–484. https://doi.org/10.1038/s41576-019-0127-1
Weedon MN, Lango H, Lindgren CM, Wallace C, Evans, DM, Mangino M, Freathy RM, Perry J. RB, Stevens S, Hall AS, Samani NJ, Shields B, Prokopenko I, Farrall M, Dominiczak A, Johnson T, Bergmann S, Beckmann, JS, Vollenweider, P, Waterworth DM, Mooser V, Palmer CNA Morris AD Ouwehand WH, Zhao JH, Li S, Loos R JF, Barroso I, Deloukas P, Sandhu MS, Wheeler E, Soranzo N, Inouye M, Wareham NJ, Caulfield M, Munroe PB, Hattersley AT, McCarthy MI, Frayling TM, Initiative, DG, Consortium TWTCC, Consortium, CG (2008) Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet 40(5):575–583. https://doi.org/10.1038/ng.121
Wilkes EH, Terfve C, Gribben JG, Saez-Rodriguez J, Cutillas PR (2015) Empirical inference of circuitry and plasticity in a kinase signaling network. Proc Natl Acad Sci U S A 112(25):7719–7724
Wray N, Wijmenga C, Sullivan P, Yang J, Visscher P (2018) Common disease is more complex than implied by the core gene omnigenic model. Cell 173:1573–1580
Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM (2010) Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42(7):565–569. https://doi.org/10.1038/ng.608
Download references
Acknowledgements
We are grateful to Simon Davis, Katie de Lange, and the anonymous reviewers (one of whom turned out to be Pierrick Bourrat) for helpful discussions and feedback. S.S. is supported by a Sir Henry Wellcome Postdoctoral Fellowship at the University of Oxford.
Author information
Authors and affiliations.
Faculty of Philosophy, University of Oxford, Oxford, UK
Weatherall Institute for Molecular Medicine, University of Oxford, Oxford, UK
Sumana Sharma
You can also search for this author in PubMed Google Scholar

Corresponding author
Correspondence to James Read .
Ethics declarations
Conflict of interest, additional information, publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Read, J., Sharma, S. Hypothesis-driven science in large-scale studies: the case of GWAS. Biol Philos 36 , 46 (2021). https://doi.org/10.1007/s10539-021-09823-0
Download citation
Received : 24 May 2021
Accepted : 08 September 2021
Published : 19 September 2021
DOI : https://doi.org/10.1007/s10539-021-09823-0
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Systems biology
- Hypothesis-driven science
- Machine learning
- Find a journal
- Publish with us
- Track your research
What is a scientific hypothesis?
It's the initial building block in the scientific method.

Hypothesis basics
What makes a hypothesis testable.
- Types of hypotheses
- Hypothesis versus theory
Additional resources
Bibliography.
A scientific hypothesis is a tentative, testable explanation for a phenomenon in the natural world. It's the initial building block in the scientific method . Many describe it as an "educated guess" based on prior knowledge and observation. While this is true, a hypothesis is more informed than a guess. While an "educated guess" suggests a random prediction based on a person's expertise, developing a hypothesis requires active observation and background research.
The basic idea of a hypothesis is that there is no predetermined outcome. For a solution to be termed a scientific hypothesis, it has to be an idea that can be supported or refuted through carefully crafted experimentation or observation. This concept, called falsifiability and testability, was advanced in the mid-20th century by Austrian-British philosopher Karl Popper in his famous book "The Logic of Scientific Discovery" (Routledge, 1959).
A key function of a hypothesis is to derive predictions about the results of future experiments and then perform those experiments to see whether they support the predictions.
A hypothesis is usually written in the form of an if-then statement, which gives a possibility (if) and explains what may happen because of the possibility (then). The statement could also include "may," according to California State University, Bakersfield .
Here are some examples of hypothesis statements:
- If garlic repels fleas, then a dog that is given garlic every day will not get fleas.
- If sugar causes cavities, then people who eat a lot of candy may be more prone to cavities.
- If ultraviolet light can damage the eyes, then maybe this light can cause blindness.
A useful hypothesis should be testable and falsifiable. That means that it should be possible to prove it wrong. A theory that can't be proved wrong is nonscientific, according to Karl Popper's 1963 book " Conjectures and Refutations ."
An example of an untestable statement is, "Dogs are better than cats." That's because the definition of "better" is vague and subjective. However, an untestable statement can be reworded to make it testable. For example, the previous statement could be changed to this: "Owning a dog is associated with higher levels of physical fitness than owning a cat." With this statement, the researcher can take measures of physical fitness from dog and cat owners and compare the two.
Types of scientific hypotheses

In an experiment, researchers generally state their hypotheses in two ways. The null hypothesis predicts that there will be no relationship between the variables tested, or no difference between the experimental groups. The alternative hypothesis predicts the opposite: that there will be a difference between the experimental groups. This is usually the hypothesis scientists are most interested in, according to the University of Miami .
For example, a null hypothesis might state, "There will be no difference in the rate of muscle growth between people who take a protein supplement and people who don't." The alternative hypothesis would state, "There will be a difference in the rate of muscle growth between people who take a protein supplement and people who don't."
If the results of the experiment show a relationship between the variables, then the null hypothesis has been rejected in favor of the alternative hypothesis, according to the book " Research Methods in Psychology " (BCcampus, 2015).
There are other ways to describe an alternative hypothesis. The alternative hypothesis above does not specify a direction of the effect, only that there will be a difference between the two groups. That type of prediction is called a two-tailed hypothesis. If a hypothesis specifies a certain direction — for example, that people who take a protein supplement will gain more muscle than people who don't — it is called a one-tailed hypothesis, according to William M. K. Trochim , a professor of Policy Analysis and Management at Cornell University.
Sometimes, errors take place during an experiment. These errors can happen in one of two ways. A type I error is when the null hypothesis is rejected when it is true. This is also known as a false positive. A type II error occurs when the null hypothesis is not rejected when it is false. This is also known as a false negative, according to the University of California, Berkeley .
A hypothesis can be rejected or modified, but it can never be proved correct 100% of the time. For example, a scientist can form a hypothesis stating that if a certain type of tomato has a gene for red pigment, that type of tomato will be red. During research, the scientist then finds that each tomato of this type is red. Though the findings confirm the hypothesis, there may be a tomato of that type somewhere in the world that isn't red. Thus, the hypothesis is true, but it may not be true 100% of the time.
Scientific theory vs. scientific hypothesis
The best hypotheses are simple. They deal with a relatively narrow set of phenomena. But theories are broader; they generally combine multiple hypotheses into a general explanation for a wide range of phenomena, according to the University of California, Berkeley . For example, a hypothesis might state, "If animals adapt to suit their environments, then birds that live on islands with lots of seeds to eat will have differently shaped beaks than birds that live on islands with lots of insects to eat." After testing many hypotheses like these, Charles Darwin formulated an overarching theory: the theory of evolution by natural selection.
"Theories are the ways that we make sense of what we observe in the natural world," Tanner said. "Theories are structures of ideas that explain and interpret facts."
- Read more about writing a hypothesis, from the American Medical Writers Association.
- Find out why a hypothesis isn't always necessary in science, from The American Biology Teacher.
- Learn about null and alternative hypotheses, from Prof. Essa on YouTube .
Encyclopedia Britannica. Scientific Hypothesis. Jan. 13, 2022. https://www.britannica.com/science/scientific-hypothesis
Karl Popper, "The Logic of Scientific Discovery," Routledge, 1959.
California State University, Bakersfield, "Formatting a testable hypothesis." https://www.csub.edu/~ddodenhoff/Bio100/Bio100sp04/formattingahypothesis.htm
Karl Popper, "Conjectures and Refutations," Routledge, 1963.
Price, P., Jhangiani, R., & Chiang, I., "Research Methods of Psychology — 2nd Canadian Edition," BCcampus, 2015.
University of Miami, "The Scientific Method" http://www.bio.miami.edu/dana/161/evolution/161app1_scimethod.pdf
William M.K. Trochim, "Research Methods Knowledge Base," https://conjointly.com/kb/hypotheses-explained/
University of California, Berkeley, "Multiple Hypothesis Testing and False Discovery Rate" https://www.stat.berkeley.edu/~hhuang/STAT141/Lecture-FDR.pdf
University of California, Berkeley, "Science at multiple levels" https://undsci.berkeley.edu/article/0_0_0/howscienceworks_19
Sign up for the Live Science daily newsletter now
Get the world’s most fascinating discoveries delivered straight to your inbox.

How much does Earth weigh?
Largest gold nugget ever found in England unearthed with faulty metal detector
Space photo of the week: James Webb telescope reveals surprising starburst in ancient galaxy
Most Popular
By Anna Gora December 27, 2023
By Anna Gora December 26, 2023
By Anna Gora December 25, 2023
By Emily Cooke December 23, 2023
By Victoria Atkinson December 22, 2023
By Anna Gora December 16, 2023
By Anna Gora December 15, 2023
By Anna Gora November 09, 2023
By Donavyn Coffey November 06, 2023
By Anna Gora October 31, 2023
By Anna Gora October 26, 2023
- 2 Polar vortex is 'spinning backwards' above Arctic after major reversal event
- 3 9,000-year-old rock art discovered among dinosaur footprints in Brazil
- 4 Where does the solar system end?
- 5 Secretive Delta IV Heavy rocket launch postponed indefinitely
- 3 The 7 most powerful supercomputers in the world right now
- 4 Fiber-optic data transfer speeds hit a rapid 301 Tbps — 1.2 million times faster than your home broadband connection
- 5 Powerful X-class solar flare slams Earth, triggering radio blackout over the Pacific Ocean
- Bipolar Disorder
- Therapy Center
- When To See a Therapist
- Types of Therapy
- Best Online Therapy
- Best Couples Therapy
- Best Family Therapy
- Managing Stress
- Sleep and Dreaming
- Understanding Emotions
- Self-Improvement
- Healthy Relationships
- Student Resources
- Personality Types
- Verywell Mind Insights
- 2023 Verywell Mind 25
- Mental Health in the Classroom
- Editorial Process
- Meet Our Review Board
- Crisis Support
How to Write a Great Hypothesis
Hypothesis Format, Examples, and Tips
Kendra Cherry, MS, is a psychosocial rehabilitation specialist, psychology educator, and author of the "Everything Psychology Book."
:max_bytes(150000):strip_icc():format(webp)/IMG_9791-89504ab694d54b66bbd72cb84ffb860e.jpg)
Amy Morin, LCSW, is a psychotherapist and international bestselling author. Her books, including "13 Things Mentally Strong People Don't Do," have been translated into more than 40 languages. Her TEDx talk, "The Secret of Becoming Mentally Strong," is one of the most viewed talks of all time.
:max_bytes(150000):strip_icc():format(webp)/VW-MIND-Amy-2b338105f1ee493f94d7e333e410fa76.jpg)
Verywell / Alex Dos Diaz
- The Scientific Method
Hypothesis Format
Falsifiability of a hypothesis, operational definitions, types of hypotheses, hypotheses examples.
- Collecting Data
Frequently Asked Questions
A hypothesis is a tentative statement about the relationship between two or more variables. It is a specific, testable prediction about what you expect to happen in a study.
One hypothesis example would be a study designed to look at the relationship between sleep deprivation and test performance might have a hypothesis that states: "This study is designed to assess the hypothesis that sleep-deprived people will perform worse on a test than individuals who are not sleep-deprived."
This article explores how a hypothesis is used in psychology research, how to write a good hypothesis, and the different types of hypotheses you might use.
The Hypothesis in the Scientific Method
In the scientific method , whether it involves research in psychology, biology, or some other area, a hypothesis represents what the researchers think will happen in an experiment. The scientific method involves the following steps:
- Forming a question
- Performing background research
- Creating a hypothesis
- Designing an experiment
- Collecting data
- Analyzing the results
- Drawing conclusions
- Communicating the results
The hypothesis is a prediction, but it involves more than a guess. Most of the time, the hypothesis begins with a question which is then explored through background research. It is only at this point that researchers begin to develop a testable hypothesis. Unless you are creating an exploratory study, your hypothesis should always explain what you expect to happen.
In a study exploring the effects of a particular drug, the hypothesis might be that researchers expect the drug to have some type of effect on the symptoms of a specific illness. In psychology, the hypothesis might focus on how a certain aspect of the environment might influence a particular behavior.
Remember, a hypothesis does not have to be correct. While the hypothesis predicts what the researchers expect to see, the goal of the research is to determine whether this guess is right or wrong. When conducting an experiment, researchers might explore a number of factors to determine which ones might contribute to the ultimate outcome.
In many cases, researchers may find that the results of an experiment do not support the original hypothesis. When writing up these results, the researchers might suggest other options that should be explored in future studies.
In many cases, researchers might draw a hypothesis from a specific theory or build on previous research. For example, prior research has shown that stress can impact the immune system. So a researcher might hypothesize: "People with high-stress levels will be more likely to contract a common cold after being exposed to the virus than people who have low-stress levels."
In other instances, researchers might look at commonly held beliefs or folk wisdom. "Birds of a feather flock together" is one example of folk wisdom that a psychologist might try to investigate. The researcher might pose a specific hypothesis that "People tend to select romantic partners who are similar to them in interests and educational level."
Elements of a Good Hypothesis
So how do you write a good hypothesis? When trying to come up with a hypothesis for your research or experiments, ask yourself the following questions:
- Is your hypothesis based on your research on a topic?
- Can your hypothesis be tested?
- Does your hypothesis include independent and dependent variables?
Before you come up with a specific hypothesis, spend some time doing background research. Once you have completed a literature review, start thinking about potential questions you still have. Pay attention to the discussion section in the journal articles you read . Many authors will suggest questions that still need to be explored.
To form a hypothesis, you should take these steps:
- Collect as many observations about a topic or problem as you can.
- Evaluate these observations and look for possible causes of the problem.
- Create a list of possible explanations that you might want to explore.
- After you have developed some possible hypotheses, think of ways that you could confirm or disprove each hypothesis through experimentation. This is known as falsifiability.
In the scientific method , falsifiability is an important part of any valid hypothesis. In order to test a claim scientifically, it must be possible that the claim could be proven false.
Students sometimes confuse the idea of falsifiability with the idea that it means that something is false, which is not the case. What falsifiability means is that if something was false, then it is possible to demonstrate that it is false.
One of the hallmarks of pseudoscience is that it makes claims that cannot be refuted or proven false.
A variable is a factor or element that can be changed and manipulated in ways that are observable and measurable. However, the researcher must also define how the variable will be manipulated and measured in the study.
For example, a researcher might operationally define the variable " test anxiety " as the results of a self-report measure of anxiety experienced during an exam. A "study habits" variable might be defined by the amount of studying that actually occurs as measured by time.
These precise descriptions are important because many things can be measured in a number of different ways. One of the basic principles of any type of scientific research is that the results must be replicable. By clearly detailing the specifics of how the variables were measured and manipulated, other researchers can better understand the results and repeat the study if needed.
Some variables are more difficult than others to define. How would you operationally define a variable such as aggression ? For obvious ethical reasons, researchers cannot create a situation in which a person behaves aggressively toward others.
In order to measure this variable, the researcher must devise a measurement that assesses aggressive behavior without harming other people. In this situation, the researcher might utilize a simulated task to measure aggressiveness.
Hypothesis Checklist
- Does your hypothesis focus on something that you can actually test?
- Does your hypothesis include both an independent and dependent variable?
- Can you manipulate the variables?
- Can your hypothesis be tested without violating ethical standards?
The hypothesis you use will depend on what you are investigating and hoping to find. Some of the main types of hypotheses that you might use include:
- Simple hypothesis : This type of hypothesis suggests that there is a relationship between one independent variable and one dependent variable.
- Complex hypothesis : This type of hypothesis suggests a relationship between three or more variables, such as two independent variables and a dependent variable.
- Null hypothesis : This hypothesis suggests no relationship exists between two or more variables.
- Alternative hypothesis : This hypothesis states the opposite of the null hypothesis.
- Statistical hypothesis : This hypothesis uses statistical analysis to evaluate a representative sample of the population and then generalizes the findings to the larger group.
- Logical hypothesis : This hypothesis assumes a relationship between variables without collecting data or evidence.
A hypothesis often follows a basic format of "If {this happens} then {this will happen}." One way to structure your hypothesis is to describe what will happen to the dependent variable if you change the independent variable .
The basic format might be: "If {these changes are made to a certain independent variable}, then we will observe {a change in a specific dependent variable}."
A few examples of simple hypotheses:
- "Students who eat breakfast will perform better on a math exam than students who do not eat breakfast."
- Complex hypothesis: "Students who experience test anxiety before an English exam will get lower scores than students who do not experience test anxiety."
- "Motorists who talk on the phone while driving will be more likely to make errors on a driving course than those who do not talk on the phone."
Examples of a complex hypothesis include:
- "People with high-sugar diets and sedentary activity levels are more likely to develop depression."
- "Younger people who are regularly exposed to green, outdoor areas have better subjective well-being than older adults who have limited exposure to green spaces."
Examples of a null hypothesis include:
- "Children who receive a new reading intervention will have scores different than students who do not receive the intervention."
- "There will be no difference in scores on a memory recall task between children and adults."
Examples of an alternative hypothesis:
- "Children who receive a new reading intervention will perform better than students who did not receive the intervention."
- "Adults will perform better on a memory task than children."
Collecting Data on Your Hypothesis
Once a researcher has formed a testable hypothesis, the next step is to select a research design and start collecting data. The research method depends largely on exactly what they are studying. There are two basic types of research methods: descriptive research and experimental research.
Descriptive Research Methods
Descriptive research such as case studies , naturalistic observations , and surveys are often used when it would be impossible or difficult to conduct an experiment . These methods are best used to describe different aspects of a behavior or psychological phenomenon.
Once a researcher has collected data using descriptive methods, a correlational study can then be used to look at how the variables are related. This type of research method might be used to investigate a hypothesis that is difficult to test experimentally.
Experimental Research Methods
Experimental methods are used to demonstrate causal relationships between variables. In an experiment, the researcher systematically manipulates a variable of interest (known as the independent variable) and measures the effect on another variable (known as the dependent variable).
Unlike correlational studies, which can only be used to determine if there is a relationship between two variables, experimental methods can be used to determine the actual nature of the relationship—whether changes in one variable actually cause another to change.
A Word From Verywell
The hypothesis is a critical part of any scientific exploration. It represents what researchers expect to find in a study or experiment. In situations where the hypothesis is unsupported by the research, the research still has value. Such research helps us better understand how different aspects of the natural world relate to one another. It also helps us develop new hypotheses that can then be tested in the future.
Some examples of how to write a hypothesis include:
- "Staying up late will lead to worse test performance the next day."
- "People who consume one apple each day will visit the doctor fewer times each year."
- "Breaking study sessions up into three 20-minute sessions will lead to better test results than a single 60-minute study session."
The four parts of a hypothesis are:
- The research question
- The independent variable (IV)
- The dependent variable (DV)
- The proposed relationship between the IV and DV
Castillo M. The scientific method: a need for something better? . AJNR Am J Neuroradiol. 2013;34(9):1669-71. doi:10.3174/ajnr.A3401
Nevid J. Psychology: Concepts and Applications. Wadworth, 2013.
By Kendra Cherry, MSEd Kendra Cherry, MS, is a psychosocial rehabilitation specialist, psychology educator, and author of the "Everything Psychology Book."
1.1 The Science of Biology
Learning objectives.
In this section, you will explore the following questions:
- What are the characteristics shared by the natural sciences?
- What are the steps of the scientific method?
Connection for AP ® courses
Biology is the science that studies living organisms and their interactions with one another and with their environment. The process of science attempts to describe and understand the nature of the universe by rational means. Science has many fields; those fields related to the physical world, including biology, are considered natural sciences. All of the natural sciences follow the laws of chemistry and physics. For example, when studying biology, you must remember living organisms obey the laws of thermodynamics while using free energy and matter from the environment to carry out life processes that are explored in later chapters, such as metabolism and reproduction.
Two types of logical reasoning are used in science: inductive reasoning and deductive reasoning. Inductive reasoning uses particular results to produce general scientific principles. Deductive reasoning uses logical thinking to predict results by applying scientific principles or practices. The scientific method is a step-by-step process that consists of: making observations, defining a problem, posing hypotheses, testing these hypotheses by designing and conducting investigations, and drawing conclusions from data and results. Scientists then communicate their results to the scientific community. Scientific theories are subject to revision as new information is collected.
The content presented in this section supports the Learning Objectives outlined in Big Idea 2 of the AP ® Biology Curriculum Framework. The Learning Objectives merge Essential Knowledge content with one or more of the seven Science Practices. These objectives provide a transparent foundation for the AP ® Biology course, along with inquiry-based laboratory experiences, instructional activities, and AP ® Exam questions.
Teacher Support
Illustrate uses of the scientific method in class. Divide students in groups of four or five and ask them to design experiments to test the existence of connections they have wondered about. Help them decide if they have a working hypothesis that can be tested and falsified. Give examples of hypotheses that are not falsifiable because they are based on subjective assessments. They are neither observable nor measurable. For example, birds like classical music is based on a subjective assessment. Ask if this hypothesis can be modified to become a testable hypothesis. Stress the need for controls and provide examples such as the use of placebos in pharmacology.
Biology is not a collection of facts to be memorized. Biological systems follow the law of physics and chemistry. Give as an example gas laws in chemistry and respiration physiology. Many students come with a 19th century view of natural sciences; each discipline is in its own sphere. Give as an example, bioinformatics which uses organism biology, chemistry, and physics to label DNA with light emitting reporter molecules (Next Generation sequencing). These molecules can then be scanned by light-sensing machinery, allowing huge amounts of information to be gathered on their DNA. Bring to their attention the fact that the analysis of these data is an application of mathematics and computer science.
For more information about next generation sequencing, check out this informative review .
What is biology? In simple terms, biology is the study of life. This is a very broad definition because the scope of biology is vast. Biologists may study anything from the microscopic or submicroscopic view of a cell to ecosystems and the whole living planet ( Figure 1.2 ). Listening to the daily news, you will quickly realize how many aspects of biology are discussed every day. For example, recent news topics include Escherichia coli ( Figure 1.3 ) outbreaks in spinach and Salmonella contamination in peanut butter. On a global scale, many researchers are committed to finding ways to protect the planet, solve environmental issues, and reduce the effects of climate change. All of these diverse endeavors are related to different facets of the discipline of biology.
The Process of Science
Biology is a science, but what exactly is science? What does the study of biology share with other scientific disciplines? Science (from the Latin scientia , meaning “knowledge”) can be defined as knowledge that covers general truths or the operation of general laws, especially when acquired and tested by the scientific method. It becomes clear from this definition that the application of the scientific method plays a major role in science. The scientific method is a method of research with defined steps that include experiments and careful observation.
The steps of the scientific method will be examined in detail later, but one of the most important aspects of this method is the testing of hypotheses by means of repeatable experiments. A hypothesis is a suggested explanation for an event, which can be tested. Although using the scientific method is inherent to science, it is inadequate in determining what science is. This is because it is relatively easy to apply the scientific method to disciplines such as physics and chemistry, but when it comes to disciplines like archaeology, psychology, and geology, the scientific method becomes less applicable as it becomes more difficult to repeat experiments.
These areas of study are still sciences, however. Consider archaeology—even though one cannot perform repeatable experiments, hypotheses may still be supported. For instance, an archaeologist can hypothesize that an ancient culture existed based on finding a piece of pottery. Further hypotheses could be made about various characteristics of this culture, and these hypotheses may be found to be correct or false through continued support or contradictions from other findings. A hypothesis may become a verified theory. A theory is a tested and confirmed explanation for observations or phenomena. Science may be better defined as fields of study that attempt to comprehend the nature of the universe.
Natural Sciences
What would you expect to see in a museum of natural sciences? Frogs? Plants? Dinosaur skeletons? Exhibits about how the brain functions? A planetarium? Gems and minerals? Or, maybe all of the above? Science includes such diverse fields as astronomy, biology, computer sciences, geology, logic, physics, chemistry, and mathematics ( Figure 1.4 ). However, those fields of science related to the physical world and its phenomena and processes are considered natural sciences . Thus, a museum of natural sciences might contain any of the items listed above.
There is no complete agreement when it comes to defining what the natural sciences include, however. For some experts, the natural sciences are astronomy, biology, chemistry, earth science, and physics. Other scholars choose to divide natural sciences into life sciences , which study living things and include biology, and physical sciences , which study nonliving matter and include astronomy, geology, physics, and chemistry. Some disciplines such as biophysics and biochemistry build on both life and physical sciences and are interdisciplinary. Natural sciences are sometimes referred to as “hard science” because they rely on the use of quantitative data; social sciences that study society and human behavior are more likely to use qualitative assessments to drive investigations and findings.
Not surprisingly, the natural science of biology has many branches or subdisciplines. Cell biologists study cell structure and function, while biologists who study anatomy investigate the structure of an entire organism. Those biologists studying physiology, however, focus on the internal functioning of an organism. Some areas of biology focus on only particular types of living things. For example, botanists explore plants, while zoologists specialize in animals.
Scientific Reasoning
One thing is common to all forms of science: an ultimate goal “to know.” Curiosity and inquiry are the driving forces for the development of science. Scientists seek to understand the world and the way it operates. To do this, they use two methods of logical thinking: inductive reasoning and deductive reasoning.
Inductive reasoning is a form of logical thinking that uses related observations to arrive at a general conclusion. This type of reasoning is common in descriptive science. A life scientist such as a biologist makes observations and records them. These data can be qualitative or quantitative, and the raw data can be supplemented with drawings, pictures, photos, or videos. From many observations, the scientist can infer conclusions (inductions) based on evidence. Inductive reasoning involves formulating generalizations inferred from careful observation and the analysis of a large amount of data. Brain studies provide an example. In this type of research, many live brains are observed while people are doing a specific activity, such as viewing images of food. The part of the brain that “lights up” during this activity is then predicted to be the part controlling the response to the selected stimulus, in this case, images of food. The “lighting up” of the various areas of the brain is caused by excess absorption of radioactive sugar derivatives by active areas of the brain. The resultant increase in radioactivity is observed by a scanner. Then, researchers can stimulate that part of the brain to see if similar responses result.
Deductive reasoning or deduction is the type of logic used in hypothesis-based science. In deductive reason, the pattern of thinking moves in the opposite direction as compared to inductive reasoning. Deductive reasoning is a form of logical thinking that uses a general principle or law to predict specific results. From those general principles, a scientist can deduce and predict the specific results that would be valid as long as the general principles are valid. Studies in climate change can illustrate this type of reasoning. For example, scientists may predict that if the climate becomes warmer in a particular region, then the distribution of plants and animals should change. These predictions have been made and tested, and many such changes have been found, such as the modification of arable areas for agriculture, with change based on temperature averages.
Both types of logical thinking are related to the two main pathways of scientific study: descriptive science and hypothesis-based science. Descriptive (or discovery) science , which is usually inductive, aims to observe, explore, and discover, while hypothesis-based science , which is usually deductive, begins with a specific question or problem and a potential answer or solution that can be tested. The boundary between these two forms of study is often blurred, and most scientific endeavors combine both approaches. The fuzzy boundary becomes apparent when thinking about how easily observation can lead to specific questions. For example, a gentleman in the 1940s observed that the burr seeds that stuck to his clothes and his dog’s fur had a tiny hook structure. On closer inspection, he discovered that the burrs’ gripping device was more reliable than a zipper. He eventually developed a company and produced the hook-and-loop fastener often used on lace-less sneakers and athletic braces. Descriptive science and hypothesis-based science are in continuous dialogue.
The Scientific Method
Biologists study the living world by posing questions about it and seeking science-based responses. This approach is common to other sciences as well and is often referred to as the scientific method. The scientific method was used even in ancient times, but it was first documented by England’s Sir Francis Bacon (1561–1626) ( Figure 1.5 ), who set up inductive methods for scientific inquiry. The scientific method is not exclusively used by biologists but can be applied to almost all fields of study as a logical, rational problem-solving method.
The scientific process typically starts with an observation (often a problem to be solved) that leads to a question. Let’s think about a simple problem that starts with an observation and apply the scientific method to solve the problem. One Monday morning, a student arrives at class and quickly discovers that the classroom is too warm. That is an observation that also describes a problem: the classroom is too warm. The student then asks a question: “Why is the classroom so warm?”
Proposing a Hypothesis
Recall that a hypothesis is a suggested explanation that can be tested. To solve a problem, several hypotheses may be proposed. For example, one hypothesis might be, “The classroom is warm because no one turned on the air conditioning.” But there could be other responses to the question, and therefore other hypotheses may be proposed. A second hypothesis might be, “The classroom is warm because there is a power failure, and so the air conditioning doesn’t work.”
Once a hypothesis has been selected, the student can make a prediction. A prediction is similar to a hypothesis but it typically has the format “If . . . then . . . .” For example, the prediction for the first hypothesis might be, “ If the student turns on the air conditioning, then the classroom will no longer be too warm.”
Testing a Hypothesis
A valid hypothesis must be testable. It should also be falsifiable , meaning that it can be disproven by experimental results. Importantly, science does not claim to “prove” anything because scientific understandings are always subject to modification with further information. This step—openness to disproving ideas—is what distinguishes sciences from non-sciences. The presence of the supernatural, for instance, is neither testable nor falsifiable. To test a hypothesis, a researcher will conduct one or more experiments designed to eliminate one or more of the hypotheses. Each experiment will have one or more variables and one or more controls. A variable is any part of the experiment that can vary or change during the experiment. The control group contains every feature of the experimental group except it is not given the manipulation that is hypothesized about. Therefore, if the results of the experimental group differ from the control group, the difference must be due to the hypothesized manipulation, rather than some outside factor. Look for the variables and controls in the examples that follow. To test the first hypothesis, the student would find out if the air conditioning is on. If the air conditioning is turned on but does not work, there should be another reason, and this hypothesis should be rejected. To test the second hypothesis, the student could check if the lights in the classroom are functional. If so, there is no power failure and this hypothesis should be rejected. Each hypothesis should be tested by carrying out appropriate experiments. Be aware that rejecting one hypothesis does not determine whether or not the other hypotheses can be accepted; it simply eliminates one hypothesis that is not valid ( see this figure ). Using the scientific method, the hypotheses that are inconsistent with experimental data are rejected.
While this “warm classroom” example is based on observational results, other hypotheses and experiments might have clearer controls. For instance, a student might attend class on Monday and realize she had difficulty concentrating on the lecture. One observation to explain this occurrence might be, “When I eat breakfast before class, I am better able to pay attention.” The student could then design an experiment with a control to test this hypothesis.
In hypothesis-based science, specific results are predicted from a general premise. This type of reasoning is called deductive reasoning: deduction proceeds from the general to the particular. But the reverse of the process is also possible: sometimes, scientists reach a general conclusion from a number of specific observations. This type of reasoning is called inductive reasoning, and it proceeds from the particular to the general. Inductive and deductive reasoning are often used in tandem to advance scientific knowledge ( see this figure ). In recent years a new approach of testing hypotheses has developed as a result of an exponential growth of data deposited in various databases. Using computer algorithms and statistical analyses of data in databases, a new field of so-called "data research" (also referred to as "in silico" research) provides new methods of data analyses and their interpretation. This will increase the demand for specialists in both biology and computer science, a promising career opportunity.
Science Practice Connection for AP® Courses
Think about it.
Almost all plants use water, carbon dioxide, and energy from the sun to make sugars. Think about what would happen to plants that don’t have sunlight as an energy source or sufficient water. What would happen to organisms that depend on those plants for their own survival?
Make a prediction about what would happen to the organisms living in a rain forest if 50% of its trees were destroyed. How would you test your prediction?
Use this example as a model to make predictions. Emphasize there is no rigid scientific method scheme. Active science is a combination of observations and measurement. Offer the example of ecology where the conventional scientific method is not always applicable because researchers cannot always set experiments in a laboratory and control all the variables.
Possible answers:
Destruction of the rain forest affects the trees, the animals which feed on the vegetation, take shelter on the trees, and large predators which feed on smaller animals. Furthermore, because the trees positively affect rain through massive evaporation and condensation of water vapor, drought follows deforestation.
Tell students a similar experiment on a grand scale may have happened in the past and introduce the next activity “What killed the dinosaurs?”
Some predictions can be made and later observations can support or disprove the prediction.
Ask, “what killed the dinosaurs?” Explain many scientists point to a massive asteroid crashing in the Yucatan peninsula in Mexico. One of the effects was the creation of smoke clouds and debris that blocked the Sun, stamped out many plants and, consequently, brought mass extinction. As is common in the scientific community, many other researchers offer divergent explanations.
Go to this site for a good example of the complexity of scientific method and scientific debate.
Visual Connection
In the example below, the scientific method is used to solve an everyday problem. Order the scientific method steps (numbered items) with the process of solving the everyday problem (lettered items). Based on the results of the experiment, is the hypothesis correct? If it is incorrect, propose some alternative hypotheses.
- The original hypothesis is correct. There is something wrong with the electrical outlet and therefore the toaster doesn’t work.
- The original hypothesis is incorrect. Alternative hypothesis includes that toaster wasn’t turned on.
- The original hypothesis is correct. The coffee maker and the toaster do not work when plugged into the outlet.
- The original hypothesis is incorrect. Alternative hypotheses includes that both coffee maker and toaster were broken.
- All flying birds and insects have wings. Birds and insects flap their wings as they move through the air. Therefore, wings enable flight.
- Insects generally survive mild winters better than harsh ones. Therefore, insect pests will become more problematic if global temperatures increase.
- Chromosomes, the carriers of DNA, are distributed evenly between the daughter cells during cell division. Therefore, each daughter cell will have the same chromosome set as the mother cell.
- Animals as diverse as humans, insects, and wolves all exhibit social behavior. Therefore, social behavior must have an evolutionary advantage.
- 1- Inductive, 2- Deductive, 3- Deductive, 4- Inductive
- 1- Deductive, 2- Inductive, 3- Deductive, 4- Inductive
- 1- Inductive, 2- Deductive, 3- Inductive, 4- Deductive
- 1- Inductive, 2-Inductive, 3- Inductive, 4- Deductive
The scientific method may seem too rigid and structured. It is important to keep in mind that, although scientists often follow this sequence, there is flexibility. Sometimes an experiment leads to conclusions that favor a change in approach; often, an experiment brings entirely new scientific questions to the puzzle. Many times, science does not operate in a linear fashion; instead, scientists continually draw inferences and make generalizations, finding patterns as their research proceeds. Scientific reasoning is more complex than the scientific method alone suggests. Notice, too, that the scientific method can be applied to solving problems that aren’t necessarily scientific in nature.
Two Types of Science: Basic Science and Applied Science
The scientific community has been debating for the last few decades about the value of different types of science. Is it valuable to pursue science for the sake of simply gaining knowledge, or does scientific knowledge only have worth if we can apply it to solving a specific problem or to bettering our lives? This question focuses on the differences between two types of science: basic science and applied science.
Basic science or “pure” science seeks to expand knowledge regardless of the short-term application of that knowledge. It is not focused on developing a product or a service of immediate public or commercial value. The immediate goal of basic science is knowledge for knowledge’s sake, though this does not mean that, in the end, it may not result in a practical application.
In contrast, applied science or “technology,” aims to use science to solve real-world problems, making it possible, for example, to improve a crop yield, find a cure for a particular disease, or save animals threatened by a natural disaster ( Figure 1.8 ). In applied science, the problem is usually defined for the researcher.
Some individuals may perceive applied science as “useful” and basic science as “useless.” A question these people might pose to a scientist advocating knowledge acquisition would be, “What for?” A careful look at the history of science, however, reveals that basic knowledge has resulted in many remarkable applications of great value. Many scientists think that a basic understanding of science is necessary before an application is developed; therefore, applied science relies on the results generated through basic science. Other scientists think that it is time to move on from basic science and instead to find solutions to actual problems. Both approaches are valid. It is true that there are problems that demand immediate attention; however, few solutions would be found without the help of the wide knowledge foundation generated through basic science.
One example of how basic and applied science can work together to solve practical problems occurred after the discovery of DNA structure led to an understanding of the molecular mechanisms governing DNA replication. Strands of DNA, unique in every human, are found in our cells, where they provide the instructions necessary for life. During DNA replication, DNA makes new copies of itself, shortly before a cell divides. Understanding the mechanisms of DNA replication enabled scientists to develop laboratory techniques that are now used to identify genetic diseases. Without basic science, it is unlikely that applied science could exist.
Another example of the link between basic and applied research is the Human Genome Project, a study in which each human chromosome was analyzed and mapped to determine the precise sequence of DNA subunits and the exact location of each gene. (The gene is the basic unit of heredity represented by a specific DNA segment that codes for a functional molecule.) Other less complex organisms have also been studied as part of this project in order to gain a better understanding of human chromosomes. The Human Genome Project ( Figure 1.9 ) relied on basic research carried out with simple organisms and, later, with the human genome. An important end goal eventually became using the data for applied research, seeking cures and early diagnoses for genetically related diseases.
While research efforts in both basic science and applied science are usually carefully planned, it is important to note that some discoveries are made by serendipity , that is, by means of a fortunate accident or a lucky surprise. Penicillin was discovered when biologist Alexander Fleming accidentally left a petri dish of Staphylococcus bacteria open. An unwanted mold grew on the dish, killing the bacteria. The mold turned out to be Penicillium , and a new antibiotic was discovered. Even in the highly organized world of science, luck—when combined with an observant, curious mind—can lead to unexpected breakthroughs.
Reporting Scientific Work
Whether scientific research is basic science or applied science, scientists must share their findings in order for other researchers to expand and build upon their discoveries. Collaboration with other scientists—when planning, conducting, and analyzing results—is important for scientific research. For this reason, important aspects of a scientist’s work are communicating with peers and disseminating results to peers. Scientists can share results by presenting them at a scientific meeting or conference, but this approach can reach only the select few who are present. Instead, most scientists present their results in peer-reviewed manuscripts that are published in scientific journals. Peer-reviewed manuscripts are scientific papers that are reviewed by a scientist’s colleagues, or peers. These colleagues are qualified individuals, often experts in the same research area, who judge whether or not the scientist’s work is suitable for publication. The process of peer review helps to ensure that the research described in a scientific paper or grant proposal is original, significant, logical, and thorough. Grant proposals, which are requests for research funding, are also subject to peer review. Scientists publish their work so other scientists can reproduce their experiments under similar or different conditions to expand on the findings.
A scientific paper is very different from creative writing. Although creativity is required to design experiments, there are fixed guidelines when it comes to presenting scientific results. First, scientific writing must be brief, concise, and accurate. A scientific paper needs to be succinct but detailed enough to allow peers to reproduce the experiments.
The scientific paper consists of several specific sections—introduction, materials and methods, results, and discussion. This structure is sometimes called the “IMRaD” format. There are usually acknowledgment and reference sections as well as an abstract (a concise summary) at the beginning of the paper. There might be additional sections depending on the type of paper and the journal where it will be published; for example, some review papers require an outline.
The introduction starts with brief, but broad, background information about what is known in the field. A good introduction also gives the rationale of the work; it justifies the work carried out and also briefly mentions the end of the paper, where the hypothesis or research question driving the research will be presented. The introduction refers to the published scientific work of others and therefore requires citations following the style of the journal. Using the work or ideas of others without proper citation is considered plagiarism .
The materials and methods section includes a complete and accurate description of the substances used, and the method and techniques used by the researchers to gather data. The description should be thorough enough to allow another researcher to repeat the experiment and obtain similar results, but it does not have to be verbose. This section will also include information on how measurements were made and what types of calculations and statistical analyses were used to examine raw data. Although the materials and methods section gives an accurate description of the experiments, it does not discuss them.
Some journals require a results section followed by a discussion section, but it is more common to combine both. If the journal does not allow the combination of both sections, the results section simply narrates the findings without any further interpretation. The results are presented by means of tables or graphs, but no duplicate information should be presented. In the discussion section, the researcher will interpret the results, describe how variables may be related, and attempt to explain the observations. It is indispensable to conduct an extensive literature search to put the results in the context of previously published scientific research. Therefore, proper citations are included in this section as well.
Finally, the conclusion section summarizes the importance of the experimental findings. While the scientific paper almost certainly answered one or more scientific questions that were stated, any good research should lead to more questions. Therefore, a well-done scientific paper leaves doors open for the researcher and others to continue and expand on the findings.
Review articles do not follow the IMRAD format because they do not present original scientific findings, or primary literature; instead, they summarize and comment on findings that were published as primary literature and typically include extensive reference sections.
As an Amazon Associate we earn from qualifying purchases.
This book may not be used in the training of large language models or otherwise be ingested into large language models or generative AI offerings without OpenStax's permission.
Want to cite, share, or modify this book? This book uses the Creative Commons Attribution License and you must attribute OpenStax.
Access for free at https://openstax.org/books/biology-ap-courses/pages/1-introduction
- Authors: Julianne Zedalis, John Eggebrecht
- Publisher/website: OpenStax
- Book title: Biology for AP® Courses
- Publication date: Mar 8, 2018
- Location: Houston, Texas
- Book URL: https://openstax.org/books/biology-ap-courses/pages/1-introduction
- Section URL: https://openstax.org/books/biology-ap-courses/pages/1-1-the-science-of-biology
© Jan 8, 2024 OpenStax. Textbook content produced by OpenStax is licensed under a Creative Commons Attribution License . The OpenStax name, OpenStax logo, OpenStax book covers, OpenStax CNX name, and OpenStax CNX logo are not subject to the Creative Commons license and may not be reproduced without the prior and express written consent of Rice University.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Sage Choice

Dogmatic modes of science
Roy s. hessels.
Experimental Psychology, Helmholtz Institute, Utrecht University, Utrecht, The Netherlands
Ignace T. C. Hooge
The scientific method has been characterised as consisting of two modes. On the one hand, there is the exploratory mode of science, where ideas are generated. On the other hand, one finds the confirmatory mode of science, where ideas are put to the test ( Tukey , 1980 ; Jaeger & Halliday , 1998 ). Various alternative labellings of this apparent dichotomy exist: data-driven versus hypothesis-driven, hypothesis-generating versus hypothesis-testing, or night science versus day science (e.g., Kell & Oliver , 2004 ; Yanai & Lercher , 2020 ). Regardless of the labelling, the dichotomy of an “idea-generating” versus an “idea-testing” mode seems pervasive in scientific thinking.
The two modes of science appear to be differentially appreciated. For example, exploratory research may carry the stink of “merely a fishing expedition” ( Kell & Oliver , 2004 ), or may be considered “weak” and yield unfavourable reviews (see the discussion of Platt [ 1964 ] in Jaeger & Halliday, 1998 ). Confirmatory research, on the other hand, seems to be considered as the holy grail in many areas of psychology (and vision science). Whether the appreciation for hypothesis-testing in psychology has been a reaction to the critique that theories in “soft areas of psychology” are “scientifically unimpressive and technologically worthless” ( Meehl, 1978 , p. 806) is an interesting question for debate. Nevertheless, the quintessential question in modern psychology is: “What is your hypothesis?” The correct answer one is expected to produce is a sentence at the level of a statistical analysis. Any other answer is wrong and yields the following response: “Ah, I see. You do exploratory research.” In Orwellian Newspeak “hypothesis” means “that which is to be decided on statistically” (cf. Yanai & Lercher , 2020 ), whereas “exploratory” means “descriptive” or even “unscientific.”
That the confirmatory mode of science is held in such high esteem is intriguing. Confirmation suggests that hypotheses or theories can be verified, a position diametrically opposed to that of, for example, Karl Popper, who claimed that theories can never be verified or confirmed, only refuted (e.g., Popper, 2002a ). Note that this cuts right into the heart of discussions on whether science can be inductive and rational or not ( Lakatos , 1978 ). It is not trivial semantics! But one does not find that a “refutatory” mode of science holds sway. Rather, refutation (or disconfirmation) is commonly avoided by the construction of ad-hoc auxiliary hypotheses when the data do not match with the theory (cf. the practice of Lakatosian defence, Meehl , 1990 ). Although sometimes frowned upon, ad-hoc hypotheses are not without merit. The observation of the planet Neptune by Galle in 1846 followed the ad-hoc hypothesis by Le Verrier and Adams (as discussed in Gershman , 2019 ): a great success for science. That ad-hoc hypotheses may also fail is evident from the hypothesised planet Vulcan by the same Le Verrier. That planet was never observed, although the discrepancy it addressed later proved to be relevant for Einstein’s theory of general relativity.
The discussion of confirmation versus refutation aside, the two-mode view of science is not merely a theoretical fancy that researchers debate about. It pervades increasingly more of the practicalities that researchers are faced with. The pre-registration movement, for example, seems to be built on this strict dichotomy. The Center for Open Science 1 writes that “Preregistration separates hypothesis-generating (exploratory) from hypothesis-testing (confirmatory) research” and this “planning improves the quality and transparency of your research.” Note the explicit normative statement here. But is a strict dichotomy of exploratory research (or data-driven or hypothesis-free) versus confirmatory research (or hypothesis-testing) sensible at all?
Is there such a thing as hypothesis-free exploration? Consider the case of a person sitting in their yard, peering at a pond through binoculars. Can we claim that this person is observing the world without hypotheses? According to Popper (2002b ), we cannot. He states: “observation is always observation in the light of theories” (p. 37). This need not be a formalised hypothesis according to the hypothetico-deductive method. Science is a human affair after all, it piggybacks on perception and cognition, which thrive through instinct, intuition, hunches, anticipation, quasi-informed guesses, and expertise (cf. Brunswik , 1955 ; Riegler , 2001 ; Chater et al. , 2018 ). Such proto-hypotheses ( Felin et al. , 2021 ) do not always lend themselves neatly to verbalisation or formalisation, or are fuzzy at best ( Rolfe , 1997 ). Thus, the decisions of where to sit, what direction to peer in, what binoculars to use, how long to wait are hardly hypothesis-free.
At the other extreme, one can ask whether hypothesis-testing is possible in the absence of exploration. Clearly, exploration in Tukey’s sense is crucial for forming a hypothesis in the first place: “Ideas come from previous exploration more often than from lightning strokes” (Tukey, 1980 , p. 23). However, devising a critical experiment to put a hypothesis to the test inevitably involves exploration. Exploration of where in the stimulus-space to measure, which parameters to use for signal processing, and so forth. This should strike a chord with the experimental scientist. Theoretically, one might be able to conceive of an experiment that can be considered as purely “hypothesis-testing.” Yet, at best it would be the hypothetical limit on a continuum between the exploratory and confirmatory modes of science.
Thus, a strict two-mode view of science is too simplistic. Nevertheless, the practical implications of such a view may be substantial, also to those who abstain from initiatives such as pre-registration. In our experience, the strict two-mode view of science permeates the thinking of e.g., institutional review boards, ethics committees, and local data archiving initiatives. The procedures derived from this strict two-mode thinking tend to take on a Kafkaesque atmosphere: The bureau of confirmatory science will see you now. It will be most pleased to guide you on your way to doing proper science.
We are happy to concede that scientific studies may be characterised as being of a more or less exploratory nature and that some studies may be characterised as clear attempts to refute or decide between scientific hypotheses. We also understand that some procedures taken up by institutional review boards, ethics committees, journals (pre-registration), and so forth, are meant to counter phenomena such as “HARKing” (the evil twin of the ad-hoc auxiliary hypothesis), “p-hacking”, blatant fraud, or to increase the replicability of science (e.g., Nosek et al. , 2012 ; Open Science Collaboration , 2015 ). Good intentions do not solely validate the means, however. What we vehemently oppose is the adoption and dogmatic use of a simplistic model of science and the scientific method that all research should adhere to. Dogma has no place in science, nor has it proved particularly effective throughout the history of science ( Feyerabend , 2010 ).
In our view, the dogmatic two-mode view of science obscures a deeper discussion—that of the goal or purpose of science. According to the influential paper by the Open Science Collaboration (2015 ) it is “that ultimate goal: truth” (p. 7). This contrasts starkly with a quote from Linschoten (1978 ):
The statement that science seeks truth is meaningless. The word “truth” either means too much or too little. It has no scientifically relatable meaning, unless truth is equivalent to relevant knowledge. Knowledge is relevant when it allows us to explain, predict, and control phenomena (p. 390). 2
If one considers hypotheses to be true or false, scientific findings to be true or false, and theories to be true or false, then a purely confirmatory way of thinking makes sense. All efforts to replicate—that is, to decide on which findings are really true—using the right statistical ( Gigerenzer , 2018 ) or methodological rituals ( Popper , 2002b ) will inevitably bring one closer to that truth. If one is less concerned with truth, and more with predicting tomorrow, then the exploratory versus confirmatory dichotomy is not all that relevant. One would rather have meaningful discussions about generalisability ( Yarkoni , 2020 ) or representativeness ( Brunswik , 1955 ; Holleman et al. , 2020 ). Anything that will yield a better prediction of tomorrow is useful, whether arrived at through Popper’s hypothetico-deductive methods, a hunch, a fishing trip, or counterinductively. According to Feyerabend ( 2010 , p. 1), “Science is an essentially anarchic enterprise: theoretical anarchism is more humanitarian and more likely to encourage progress than its law-and-order alternatives.” Science needs no dogma.
Acknowledgements
The authors thank Andrea van Doorn and Jan Koenderink for inspiring discussions and helpful comments.
1. https://www.cos.io/initiatives/prereg , accessed 14 May 2021.
2. In the original Dutch: “De uitspraak dat wetenschap waarheid zoekt, is zinloos. Het woord ‘waarheid’ betekent te veel of te weinig. Het geeft geen wetenschappelijk verbindbare betekenis, tenzij waarheid gelijkluidend is met relevante kennis. Kennis is relevant wanneer ze ons in staat stelt verschijnselen te verklaren, te voorspellen, en te beheersen.”
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship and/or publication of this article.
Funding: The authors received no financial support for the research, authorship and/or publication of this article.
ORCID iD: Roy S. Hessels https://orcid.org/0000-0002-4907-1067
Supplemental material
Supplemental material for this article is available online.

VIDEO
COMMENTS
Developing a hypothesis (with example) Step 1. Ask a question. Writing a hypothesis begins with a research question that you want to answer. The question should be focused, specific, and researchable within the constraints of your project. Example: Research question.
Hypothesis Driven Science • Driven by deductive reasoning. • Guided by a hypothesis (a tentative answer to a question) based on an observation. • If a hypothesis is correct, and we test it, then we can expect a particular outcome. • Experiments designed to test hypotheses must be controlled experiments. Inductive vs. Deductive Reasoning
Observations lead to questions, questions lead to forming a hypothesis as a possible answer to those questions, and then the hypothesis is tested. Thus, descriptive science and hypothesis-based science are in continuous dialogue. Hypothesis Testing. Biologists study the living world by posing questions about it and seeking science-based responses.
hypothesis. science. scientific hypothesis, an idea that proposes a tentative explanation about a phenomenon or a narrow set of phenomena observed in the natural world. The two primary features of a scientific hypothesis are falsifiability and testability, which are reflected in an "If…then" statement summarizing the idea and in the ...
In a hypothesis-driven research, specifications of methodology help the grant reviewers to differentiate good science from bad science, and thus, hypothesis-driven research is the most funded research. "Hypotheses aren't simply useful tools in some potentially outmoded vision of science; they are the whole point."
An example is a linear metabolic pathway, in which feedback inhibition is essentially always exerted on the first step ... now what is the hypothesis? The complementary roles of inductive and hypothesis-driven science in the post-genomic era. Bioessays. 2004;26(1):99-105. pmid:14696046 . View Article PubMed/NCBI Google Scholar 35.
Make observations. Formulate a hypothesis. Design an experiment to test the hypothesis. State the indicators to evaluate if the experiment has succeeded. Conduct the experiment. Evaluate the results of the experiment. Accept or reject the hypothesis. If necessary, make and test a new hypothesis.
The scientific method. At the core of biology and other sciences lies a problem-solving approach called the scientific method. The scientific method has five basic steps, plus one feedback step: Make an observation. Ask a question. Form a hypothesis, or testable explanation. Make a prediction based on the hypothesis.
For example, hypothesis-driven approaches are particularly helpful for choosing between alternative mechanisms that could explain an observed phenomenon (e.g., through a controlled experiment), but they are much less helpful for mapping out new areas of inquiry (e.g., the sequence of the human genome), identifying important relationships among ...
Ratti characterises such studies, and their connections with more orthodox mechanistic hypothesis-driven science, as involving three steps: 1. ... Many applications of the framework presented in the introduction—perform genome-wide screens based on a general hypothesis (for example, 'a gene/process is responsible for a disease'), and on ...
A scientific hypothesis is a tentative, testable explanation for a phenomenon in the natural world. It's the initial building block in the scientific method. Many describe it as an "educated guess ...
Science progresses in a dualistic fashion. You can either generate a new hypothesis out of existing data and conduct science in a data-driven way, or generate new data for an existing hypothesis and conduct science in a hypothesis-driven way. For instance, when Kepler was looking at the astronomical data sets to come up with his laws of planetary motion, he was doing data-driven science.
Simple hypothesis: This type of hypothesis suggests that there is a relationship between one independent variable and one dependent variable.; Complex hypothesis: This type of hypothesis suggests a relationship between three or more variables, such as two independent variables and a dependent variable.; Null hypothesis: This hypothesis suggests no relationship exists between two or more variables.
A hypothesis (plural hypotheses) is a proposed explanation for an observation. The definition depends on the subject. In science, a hypothesis is part of the scientific method. It is a prediction or explanation that is tested by an experiment. Observations and experiments may disprove a scientific hypothesis, but can never entirely prove one.
Scientific Hypothesis Examples . Hypothesis: All forks have three tines. This would be disproven if you find any fork with a different number of tines. Hypothesis: There is no relationship between smoking and lung cancer.While it is difficult to establish cause and effect in health issues, you can apply statistics to data to discredit or support this hypothesis.
A hypothesis is a tentative, testable answer to a scientific question. Once a scientist has a scientific question she is interested in, the scientist reads up to find out what is already known on the topic. Then she uses that information to form a tentative answer to her scientific question. Sometimes people refer to the tentative answer as "an ...
In hypothesis-based science, specific results are predicted from a general premise. This type of reasoning is called deductive reasoning: deduction proceeds from the general to the particular. But the reverse of the process is also possible: sometimes, scientists reach a general conclusion from a number of specific observations.
Terms used to describe non-hypothesis-driven research are 'descriptive research,' in which information is collected without a particular question in mind, and 'discovery science,' where large volumes of experimental data are analyzed with the goal of finding new patterns or correlations. ... Example 1: "Disease X results from the ...
This is then refuted, confirmed or reframed based on evidence. The following are illustrative examples of a hypothesis.Plants will grow faster in blue light as compared to red or green light.Regular watering can desalinate soil in a pot.Local air quality is better on weekends and holidays.Tennis balls bounce higher when they are cold.There is ...
Here we discuss the two major types of how to do science: discovery-based science, and hypothesis-based science.Table of Contents:01:11 - 01:33 - Discover re...
For example, exploratory research may carry the stink of "merely a fishing expedition" (Kell & Oliver, ... Here is the evidence, now what is the hypothesis? the complementary roles of inductive and hypothesis-driven science in the post-genomic era. BioEssays, 26 (1), 99-105. 10.1002/bies.10385 [Google Scholar] Lakatos I. (1978). ...
Which of the following is an example of hypothesis-driven science? testing whether or not dead batteries are preventing a TV remote from working. Which of the following lists the steps of the scientific method in their proper order? observation, question, hypothesis, prediction, experiment.
Which of the following is an example of hypothesis-driven science? A. Surveying the distribution of various majors at your school B. Sequencing the human genome C. Testing whether or not dead batteries are preventing a TV remote from working D. Observing the activities of a pride of lions