Computational and Systems Biology Ph.D. Program (CSB)
Principal Investigator Christopher Burge
Co-investigators Joel Voldman , Forest White , Jacob White , Ron Weiss , Alan Grossman , Amy Keating , B L Whang , Eric Alm , Mark Bathe , Ernest Fraenkel , Mehmet Fatih Yanik
Project Website http://csbi.mit.edu/education/phd.html
The Computational and Systems Biology (CSB) Ph.D. Program prepares students to become independent, interdisciplinary researchers in post-genomic biology and related fields. There is a strong focus on quantitative methods and modeling, experimental design, and device development. This unique program integrates MIT's world-class research and educational opportunities in biology, engineering, and computer science.
The emerging field of systems biology represents an integration of concepts and ideas from the biological sciences, engineering disciplines, and computer science. Recent advances in biology, including the Human Genome Project and massively parallel approaches to probing biological samples, have created new opportunities to understand biological problems from a systems perspective. Systems modeling and design are well established in engineering disciplines but are relatively new to biology. Microsystems has been fundamental to advances in electronics and computing, but now has the potential to revolutionize biology as well. Advances in systems biology will require multidisciplinary teams to apply principles and tools from engineering and computer science to solve problems in biology and medicine.
To provide training in this emerging field, MIT offers a Ph.D. Program in Computational and Systems Biology (the CSB Ph.D. Program). Spanning the School of Engineering and the School of Science, the program integrates coursework and research opportunities in biology, engineering, mathematics, microsystems, and computer science with interdisciplinary courses in computational and systems biology developed for this program. Graduates of the program will be uniquely prepared to develop original methods, make discoveries, and establish new paradigms. They will also be well-positioned to assume critical leadership roles in academia and industry, where this new research area is of growing importance.


Office Directory
Csb: computational systems biology phd program.

Curriculum & Core Subjects
The CSB Ph.D. curriculum has two components: the core subjects and advanced electives. Core subjects provide foundational knowledge of both biology and computational biology. Advanced electives are chosen by each student to generate a customized program of study, in close consultation with members of the CSB Ph.D. Graduate Committee and the student's thesis advisor. The goal is to allow students broad latitude in defining their individual area of interest, but at the same time to provide oversight and guidance to ensure that they receive rigorous and thorough training.
Core Subjects
The core curriculum consists of three classroom subjects plus a set of three two-month rotations in different research groups. The classroom subjects fall into three areas:
Topics in Computational and Systems Biology (One Subject): All first-year students in the program are required to participate in this literature-based exploration of current research frontiers and paradigms. Papers for discussion are selected from a broad range of topics in computational and systems biology, with an emphasis on the integration of experimental and computational approaches to understanding complex biological systems. This subject is limited to students in the CSB Ph.D. Program in order to build a strong community among the class. It is the only subject in the program with such a limitation.
CSB.100 Topics in Computational and Systems Biology
Modern Biology (One Subject):
A semester of modern graduate-level biology at MIT strengthens the biology base of all students in the program. Subjects in molecular biology, neurobiology, biochemistry, or genetics fulfill this requirement. The particular course taken by each student will depend on his or her background and will be determined in consultation with members of the CSB Ph.D. Graduate Committee. Subjects that can fulfill the biology requirement for the CSB Ph.D. degree include: (choose one)
- Principles of Biochemical Analysis (7.51)
- Genetics for Graduate Students (7.52)
- Molecular Biology (7.58)
- Eukaryotic Cell Biology: Principles and Practice (7.61/20.561J)
- Immunology (7.63)
- Molecular and Cellular Neuroscience Core II (7.68/9.013J)
Computational Biology (One Subject):
1. 6.8700/HST.507 J Advanced Computational Biology: Genomes, Networks, Evolution . This course additionally examines recent publications in the areas covered, with research-style assignments. A more substantial final project is expected, which can lead to a thesis and publication,
2. 7.81/8.591 J Systems Biology This graduate-level course explores more in-depth cellular and population-level systems with an emphasis on synthetic biology, modeling of genetic networks, cell-cell interactions, and evolutionary dynamics.
3. 20.490 Computational Systems Biology: Deep Learning in the Life Sciences Presents innovative approaches to computational problems in the life sciences, focusing on deep learning-based approaches with comparisons to conventional methods. Topics include protein-DNA interaction, chromatin accessibility, regulatory variant interpretation, medical image understanding, medical record understanding, therapeutic design, and experiment design (the choice and interpretation of interventions). Focuses on machine learning model selection, robustness, and interpretation. Teams complete a multidisciplinary final research project using TensorFlow or other framework. Provides a comprehensive introduction to each life sciences problem, but relies upon students understanding probabilistic problem formulations. Students taking graduate version complete additional assignments.
4. Both a and b below:
a. 6.C51Modeling with Machine Learning: from Algorithms to Applications focuses on modeling with machine learning methods with an eye towards applications in engineering and sciences. Introduction to modern machine learning methods, from supervised to unsupervised models, with an emphasis on newer neural apporaches. Emphasis on the understanding of how and why the methods work from the point of view of modeling, and when they are applicable. Unsing concrete examples, covers formulation of machine learning tasks, adapting and extending methods to given problems, and how the methods can and should be evaluated. Students taking graduate version complete additonal assignments. Students cannot receive credit without simultaneous completion of a 6-unit disciplinary module. Enrollment may be limited.
b. 20.C51/3.C51/10.C51J Machine Learning for Molecular Engineering Building on core material in 6.C51, provides an introduction to the use of machine learning to solve problems arising int he science and engineering of biology, chemistry, and materials. Equips students to design and implement achine learning approaches to challenges such as analysis of omics (genomics, transcriptomics, proteomics, etc.) microscopy, spectroscopy, or crystallography data and design of new molecules and materials such as drugs, catalysts, polymer, alloys, ceramics, and proteins. Students taking graduat version complete addiitonal assignments. Students cannot receive credit with simultaneous completion of 6.CS1.
6.C51 & 20.C51 MUST BE TAKEN TOGETHER IN THE SAME SEMESTER
- Research Areas
- Selected Publications
- Accessibility
MIT Computational Biology Group - Positions Available
Feb 2023: We have an opening for a joint position with Marinka Zitnik's group for Machine Learning for Medicine and Science at Harvard, focused on algorithm design for use in applications including biomedical discovery, drug discovery and development, and therapeutics. We are especially looking for candidates with a background in machine learning, explainable AI/ML, computational healthcare, and network science.
- On the computational side, we seek applicants with strong experience in computer science (programming, statistics, machine learning, algorithms, software engineering) and computational biology (genomics, epigenomics, regulatory genomics, statistical genetics, disease genomics). Ideal applicants should have a strong theoretic background in method development, and also practical experience in large-scale data analysis. In addition to openings within our group, we currently also have an opening for a joint position with Marinka Zitnik's group at Harvard.
- On the experimental side, experience in next-gen sequencing assays (including ChIP-Seq, DNase-Seq, ATAC-Seq, mC-Seq), capture technologies, CRISRP-Cas9, mammalian cell culature, selection and screening assays, assay development, optimization, adaptation, and multiplexing.
- The disease areas that we're focusing on include metabolic, neurodegenerative, psychiatric, and immune disorders and cancer , and domain expertise in obesity, type 2 diabetes, Alzheimer's Disease, immune cells, and cancer are welcome.
- Regardless of your specific background and project, you will end up being exposed to a broad range of computational and experimental aspects of computational biology, in a highly interdisciplinary team , and you'll have the opportunity to participate in all aspects of experiment design, assay development, sample coordination, method development, software implementation, data analysis, and experimental validation.
- Lastly, we're also looking for a project manager to help coordinate lab activities, includingg large-scale data production, technology development, next-generation sequencing, and coordination between our computational and experimental teams.
- Please fill out the application form below or at the single-page form .
- Email [email protected] a current CV with research background and publications list, a 1-2 page research statement with the specific areas you want to work on, how they fit with our group, and how they build upon your previous work, a slide deck from a recent presentation of your work, and recording of you presenting the work (approx. 35-40 minutes).
- Ask for 3 recommendation letters from your previous research supervisors and collaborators to be sent directly to [email protected] .
- If you don't hear back within a week, please re-send your email.
Ph.D. applicants: If you are interested in applying to MIT, please consider the Computer Science (EECS) and Computational and Systems Biology (CSBi) and Biological Engineering and Health Science and Technology graduate programs. Admitted graduate students are invited for on-campus visits to MIT, which is a great opportunity to meet with faculty members. You can indicate your interest in working with Prof. Kellis in your application, but admissions are done centrally, and not with a specific professor. Those contacts are established during visit weekend.
MIT graduate students: If you are already admitted to MIT, you should contact Manolis Kellis directly and set up a meeting to talk research. Reach out to [email protected] if you don't hear back immediately, ring me at 617-253-2419, or just stop by (32D-524). If you do not have prior experience in computational biology, please take 6.047/6.878 "Computational Biology: Genomes, Networks, Evolution". A strong background in algorithms or machine learning is a big plus, and some background in biology is encouraged though not required.
Undergraduate students: To be most productive in the group, we recommend that you first take 6.047/6.878 "Computational Biology: Genomes, Networks, Evolution", as it will give you the breadth needed for selecting a project, and hands-on experience in a final project that can evolve into a UROP project during the Spring term or Summer. We also encourage our UROP students to continue their research experience through a UAP or MEng project.
Application form:
(1) integrative analysis of genomic and epigenomic datasets in human.
Our group is working closely with ENCODE and the Epigenome Roadmap to integrate epigenomic information from many chromatin marks and other epigenetic modifications in multiple cell lines in order to discover and interpret chromatin states, understand their biological properties and function, and study their dynamics across multiple cell types.
Project involves computational analysis of large-scale ChIP-seq datasets (Chromatin Immunoprecipitation followed by Solexa/Illumina sequencing) to identify DNA elements associated with changes in chromatin state across different cell types and during cell differentiation. Strong background in computer science and computational biology needed. Background in epigenetics highly encouraged. Applicant will interact with the computational biology group, and also our collaborators at the Broad Institute, and across the ENCODE and Epigenome Roadmap consortia.
(2) INTERPRETING HUMAN DISEASE ASSOCIATION STUDIES
(3) integrative analysis of the human encode project, (4) regulatory genomics in human and model organisms, (5) non-coding rnas and chromatin, (6) mammalian and human comparative and population genomics, (7) genome evolution and phylogenomics.
We are interested in the principles underlying gene and genome evolution, and in particular the forces of mutation and selection acting at the gene level and at the species level. We construct probabilistic models of gene tree and species tree evolution, and seek to distinguish gene duplication, horizontal gene transfer, incomplete lineage sorting, in order to understand the sources and remedies for gene tree incongruence. We are also using such methods and models to refine species phylogenies in the eukaryotic world in the context of the tree of life project with the Eric Alm lab. Lastly, we seek to understand the forces governing the evolution of regulatory motifs and the re-wiring of regulatory networks. Applicants should have extensive comparative genomics and phylogenomics experience and relevant publications in the field.
OTHER PROJECTS
Graduate Programs
Computational biology.
The Center for Computational Molecular Biology (CCMB) offers Ph.D. degrees in Computational Biology to train the next generation of scientists to perform cutting edge research in the multidisciplinary field of Computational Biology.
During the course of their Ph.D. studies students will develop and apply novel computational, mathematical , and statistical techniques to problems in the life sciences. Students in this program must achieve mastery in three areas - computational science, molecular biology, and probability and statistical inference - through a common core of studies that spans and integrates these areas.
The Ph.D. program in Computational Biology draws on course offerings from the disciplines of the Center’s Core faculty members. These areas are Applied Mathematics, Computer Science, the Division of Biology and Medicine, the Center for Biomedical Informatics, and the School of Public Health. Our faculty and Director of Graduate Studies work with each student to develop the best plan of coursework and research rotations to meet the student’s goals in their research focus and satisfy the University’s requirements for graduation.
Applicants should state a preference for at least one of these areas in their personal statement or elsewhere in their application. In addition, students interested in the intersection of Applied Mathematics and Computational Biology are encouraged to apply directly to the Applied Mathematics Ph.D. program , and also to contact relevant CCMB faculty members .
Our Ph.D. program assumes the following prerequisites: mathematics through intermediate calculus, linear algebra and discrete mathematics, demonstrated programming skill, and at least one undergraduate course in chemistry and in molecular biology. Exceptional strengths in one area may compensate for limited background in other areas, but some proficiency across the disciplines must be evident for admission.
Additional Resources
CCMB computing resources include a set of multiprocessor computer clusters and data storage servers with 392 processors. The CCMB Cluster is the largest dedicated computing system on campus for computational biology and bioinformatics applications. See also answers to frequently asked questions .
Application Information
Application requirements, gre subject:.
Not required
GRE General:
Personal statement:.
Applicants will be asked a series of short form questions regarding their interest in computational biology, their research experiences, and their goals for the future. 1) Describe the life experiences that inspired you to pursue a career in science. 2) Describe at least one research experience you have had that prepared you intellectually/ scientifically for a career in computational biology. 3) Explain at least one challenge you have overcome in life or research to pursue a scientific career and what you have learned from this experience. 4) Discuss any broader impacts that you have had on your community (e.g. family, educational institution, or broader community). 5) Why would you like to pursue your PhD in the Brown CCMB program? (Include at least two faculty members who you would like to work with at Brown and why.)
Dates/Deadlines
Application deadline, completion requirements.
Six graduate–level courses, two eight–week laboratory rotations, preliminary research presentation, dissertation, oral defense
Contact and Location
Center for computational molecular biology, location address, mailing address.
- Program Faculty
- Program Handbook
- Graduate School Handbook
Support Biology
Dei council and dei faculty committee, biology diversity community, mit biology catalyst symposium, honors and awards, employment opportunities, faculty and research, current faculty, in memoriam, areas of research, biochemistry, biophysics, and structural biology, cancer biology, cell biology, computational biology, human disease, microbiology, neurobiology, stem cell and developmental biology, core facilities, video gallery, faculty resources, undergraduate, why biology, undergraduate testimonials, major/minor requirements, general institute requirement, advanced standing exam, transfer credit, current students, subject offerings, research opportunities, biology undergraduate student association, career development, why mit biology, diversity in the graduate program, nih training grant, career outcomes, graduate testimonials, prospective students, application process, interdisciplinary and joint degree programs, living in cambridge, graduate manual: key program info, graduate teaching, career development resources, biology graduate student council, biopals program, postdoctoral, life as a postdoc, postdoc associations, postdoc testimonials, workshops for mit biology postdocs entering the academic job market, responsible conduct of research, postdoc resources, non-mit undergraduates, bernard s. and sophie g. gould mit summer research program in biology (bsg-msrp-bio), bsg-msrp-bio gould fellows, quantitative methods workshop, high school students and teachers, summer workshop for teachers, mit field trips, leah knox scholars program, additional resources, mitx biology, biogenesis podcast, biology newsletter, department calendar, ehs and facilities, graduate manual, resources for md/phd students, preliminary exam guidelines, thesis committee meetings, guidelines for graduating, mentoring students and early-career scientists, remembering stephen goldman (1962 – 2022).
Our world-renowned faculty include 3 Nobel laureates; 29 members of the National Academy of Sciences; 11 Howard Hughes Medical Institute (HHMI) investigators; and 4 recipients of the National Medal of Science.

Tania A. Baker
Tania Baker’s current research explores mechanisms and regulation of enzyme-catalyzed protein unfolding, ATP-dependent protein degradation, and remodeling of the proteome during cellular stress responses.

David Bartel
David Bartel studies molecular pathways that regulate eukaryotic gene expression by affecting the stability or translation of mRNAs.

Facundo Batista
Facundo Batista studies fundamental lymphocyte biology to drive the development of the next generation of vaccines and therapeutics.

Stephen Bell
Stephen Bell probes the cellular machinery that replicates and maintains animal cell chromosomes.

Laurie A. Boyer
Co-Undergrad Officer
Laurie A. Boyer investigates the gene regulatory mechanisms that drive heart development and regeneration using embryonic stem cells and mouse models.

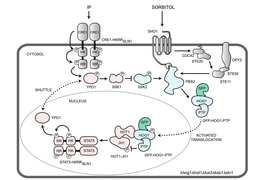
Christopher Burge
Christopher Burge applies a combination of experimental and computational approaches to understand the regulatory codes underlying pre-mRNA splicing and other types of post-transcriptional gene regulation.

Eliezer Calo
Eliezer Calo studies how cells build ribosomes and how dysfunction in ribosome biogenesis and function leads to tissue-specific developmental disorders and cancer.

Lindsay Case
Lindsay Case studies how molecules are concentrated and organized at the plasma membrane to regulate transmembrane signaling.

Iain M. Cheeseman
Associate Dept. Head
Iain Cheeseman analyzes the process by which cells duplicate, focusing on how the molecular machinery that segregates the chromosomes is rewired across diverse physiological contexts.

Jianzhu Chen
Jianzhu Chen studies the immune system, harnessing the body’s defense force to explore treatment and prevention for cancer, as well as metabolic and infectious diseases.

Yiyin Erin Chen
Erin Chen studies how the microbes in our bodies educate our immune systems, in order to engineer microbial therapeutics for human disease.

Sallie (Penny) W. Chisholm
Sallie (Penny) W. Chisholm studies the biology, ecology, and evolution of the single most abundant marine phytoplankton species in order to understand the forces that shape microbial ecosystems.

Olivia Corradin
Olivia Corradin investigates the genetic and epigenetic changes in gene regulatory elements that influence human disease.

Joseph (Joey) Davis
Joey Davis investigates how cells maintain a delicate internal balance of assembling and dismantling their own machinery — in particular, assemblages of many molecules known as macromolecular complexes.

Catherine Drennan
Catherine Drennan takes “snapshots” of metalloenzymes using crystallography and/or cryo-electron microscopy.

Gerald R. Fink
Gerald R. Fink investigates how fungal pathogens invade the body, evade the immune system, and establish an infection.

Mary Gehring
Graduate Officer
Mary Gehring researches epigenetic mechanisms of gene regulation in plants.

Alan D. Grossman
Alan Grossman studies mechanisms and regulation of DNA replication, gene expression, and horizontal gene transfer in bacteria.

Leonard P. Guarente
Leonard P. Guarente looks at mammal, mouse, and human brains to understand the genetic underpinning of aging and age-related diseases like Alzheimer’s.

Michael T. Hemann
Michael T. Hemann uses mouse models to combat cancers resistant to chemotherapy.

Whitney Henry
Whitney Henry studies ferroptosis in human health and disease with a focus on cancer.

H. Robert Horvitz
H. Robert Horvitz analyzes the roles of genes in animal development and behavior, gaining insight into human disease.

David Housman
David Housman studies the biological underpinnings of diseases like Huntington’s, cancer, and cardiovascular disease.

Siniša Hrvatin
Siniša Hrvatin studies states of stasis, such as mammalian torpor and hibernation, as a means to harness the potential of these biological adaptations to advance medicine.

Richard O. Hynes
Richard O. Hynes investigates the network of proteins surrounding cells to understand its roles in the spread of cancer throughout the body.

Barbara Imperiali
Barbara Imperiali studies the biogenesis and myriad functions of glycoconjugates in human health and disease.

Tyler Jacks
Tyler Jacks is interested in the genetic events contributing to the development of cancer, and his group has created a series of mouse strains engineered to carry mutations in genes known to be involved in human cancers.

Rudolf Jaenisch
Rudolf Jaenisch uses pluripotent cells (ES and iPS cells) to study the genetic and epigenetic basis of human diseases such as Parkinson’s, Alzheimer’s, autism and cancer.

Ankur Jain investigates the role of RNA self-assembly in cellular organization and neurodegenerative disease.

Chris A. Kaiser
Before closing his lab, Chris A. Kaiser analyzed protein folding and trafficking in cells.

Amy E. Keating
Department Head
Amy E. Keating determines how proteins make specific interactions with one another and designs new, synthetic protein-protein interactions.

Kristin Knouse
Kristin Knouse seeks to understand and modulate organ injury and repair by innovating tools for experimentation directly within living organisms.

Sally Kornbluth
President of MIT
Sally Kornbluth is President of MIT.

Monty Krieger
Monty Krieger studies cell surface receptors and cholesterol and their impact on normal physiology and diseases, such as heart disease and infertility.

Rebecca Lamason
Rebecca Lamason investigates what happens when cellular functions are hijacked by unwanted interlopers: namely, the bacteria that engender diseases like spotted fever and meningitis.

Eric S. Lander
Eric S. Lander is interested in every aspect of the human genome and its application to medicine.

Michael T. Laub
Michael T. Laub explores how bacterial cells process information and regulate their own growth and proliferation, as well as how these information-processing capabilities have evolved.

Douglas Lauffenburger
Douglas Lauffenburger fosters the interface of bioengineering, quantitative cell biology, and systems biology to determine fundamental aspects of cell dysregulation — identifying and testing new therapeutic ideas.

Jacqueline Lees
Jacqueline Lees develops mouse and zebrafish models, identifying the molecular pathways leading to tumor formation.

Ruth Lehmann
Ruth Lehmann studies the biological origins of germ cells, and how they transmit the potential to build a completely new organism to their offspring.

Daniel Lew uses fungal model systems to ask how cells orient their activities in space, including oriented growth, cell wall remodeling, and organelle segregation.

Gene-Wei Li
Gene-Wei Li investigates how quantitative information regarding precise proteome composition is encoded in and extracted from bacterial genomes.

Pulin Li is interested in quantitatively understanding how genetic circuits create multicellular behavior in both natural and synthetically engineered systems.

Troy Littleton
Troy Littleton is interested in how neuronal connections form and function, and how neurological disease disrupts synaptic communication.

Harvey F. Lodish
Before closing his lab, Harvey F. Lodish studied the development of red blood cells and the use of modified red cells for the introduction of novel therapeutics into the human body, as well as the development of brown and white fat cells.

Sebastian Lourido
Sebastian Lourido exposes parasite vulnerabilities and harnesses them to treat infectious disease.

Adam C. Martin
Adam C. Martin studies molecular mechanisms that underlie tissue form and function.

Hernandez Moura Silva
Hernandez Moura Silva seeks to understand how the immune system supports tissue physiology to unveil new approaches to treat human diseases.

Elly Nedivi
Elly Nedivi studies the mechanisms underlying brain circuit plasticity — characterizing the genes and proteins involved, as well as visualizing synaptic and neuronal remodeling in the living mouse brain.

Sergey Ovchinnikov
Sergey Ovchinnikov studies protein structure and evolution at environmental, organismal, genomic, structural, and molecular scales.

David C. Page
David C. Page examines the genetic differences between males and females — and how these play out in disease, development, and evolution.

Sara Prescott
Sara Prescott investigates how sensory inputs from within the body control mammalian physiology and behavior.

Peter Reddien
Peter Reddien works to unravel one of the greatest mysteries in biology — how organisms regenerate missing body parts.

Alison E. Ringel
Alison E. Ringel seeks to understand the molecular adaptations that enable immune cells to function and survive within unfavorable environments.

Francisco J. Sánchez-Rivera
Francisco J. Sánchez-Rivera aims to understand how genetic variation shapes normal physiology and disease, with a focus on cancer.

Robert T. Sauer
Bob Sauer studies intracellular proteolytic machines responsible for protein-quality control and homeostasis.

Thomas U. Schwartz
Thomas U. Schwartz investigates communication across biological membranes, using structural, biochemical, and genetic tools.

Anthony J. Sinskey
Anthony J. Sinskey explores the principles of metabolic engineering in both bacteria and plants.

Yadira Soto-Feliciano
Yadira Soto-Feliciano studies chromatin and epigenetic regulation in normal development and cancer.

Stefani Spranger
Stefani Spranger studies how the body’s immune system interacts with growing tumors to harness the immune response to fight cancer.

Susumu Tonegawa
Susumu Tonegawa investigates the biological underpinnings of learning and memory in rodents.

Matthew Vander Heiden
Matthew Vander Heiden is interested in the role that cell metabolism plays in mammalian physiology, with a focus on cancer.

Seychelle M. Vos
Seychelle M. Vos investigates how genome organization and gene expression are physically coupled across molecular scales.

Graham C. Walker
Graham C. Walker studies DNA repair, mutagenesis, and cellular responses to DNA damage, as well as the symbiotic relationship between legumes and nitrogen-fixing bacteria.

Bruce Walker
Bruce Walker investigates cellular immune responses in chronic human viral infections, with a particular focus on HIV immunology and vaccine development.

Robert A. Weinberg
Robert A. Weinberg studies how cancer spreads, what gives cancer stem-cells their unique qualities, and the molecular players involved in the formation of cancer stem cells and metastases.


Brandon Weissbourd
Brady Weissbourd uses jellyfish to study nervous system evolution, development, regeneration, and function.

Jonathan Weissman
Jonathan Weissman investigates how proteins fold into their correct shape and how misfolding impacts disease and normal physiology, while building innovative tools for exploring the organizational principles of biological systems.

Matthew A. Wilson
Matthew Wilson studies rodent learning and memory by recording and manipulating the activity of neurons during behavior and sleep.

Harikesh S. Wong
Harikesh S. Wong studies how cells assemble and communicate to control immune responses in tissues.

Michael B. Yaffe
Michael B. Yaffe studies the chain of reactions that controls a cell’s response to stress, cell injury, and DNA damage.

Yukiko Yamashita
Yukiko Yamashita studies two fundamental aspects of multicellular organisms: how cell fates are diversified via asymmetric cell division, and how genetic information is transmitted through generations via the germline.

Omer H. Yilmaz
Omer H. Yilmaz explores the impact of dietary interventions on stem cells, the immune system, and cancer within the intestine.

Richard A. Young
Richard A. Young explores how and why gene expression differs in healthy versus diseased cells.
Emeritus Faculty

Martha Constantine-Paton
Professor Emerita
Before closing her lab, Martha Constantine-Paton used a combination of classical and modern genetic tools in mice to study the contributions of specific brain regions to normal behavior.

Malcolm Gefter
Professor Emeritus

Frank Gertler
Before closing his lab, Frank B. Gertler considered the role of cell shape and movement in developmental defects and diseases.

Nancy Hopkins
Before closing her lab, Nancy Hopkins worked on the genetics of mouse RNA tumor viruses; on the genetics of early vertebrate development using zebrafish; and on the fish as a cancer model.

Jonathan A. King
Before closing his lab, Jonathan A. King studied what happens when proteins do not fold properly — leading to conditions like cataracts. He currently works to protect the conditions needed to support biomedical research.

Terry Orr-Weaver
Before closing her lab, Terry Orr-Weaver probed the incredibly complex and coordinated process of development from egg to fertilized embryo and ultimately adult.

Mary-Lou Pardue
Before closing her lab, Mary-Lou Pardue studied fruit fly chromosomes to better understand chromosome replication, cell division, and related cellular structures.

William (Chip) Quinn
Before closing his lab, William Quinn analyzed the molecular and genetic underpinnings of learning and memory in fruit flies before retiring.

Uttam RajBhandary
Before closing his lab, Uttam RajBhandary studied interactions between RNAs and proteins, focusing on gene expression and gene regulation.

Phillips Robbins
Professor of Biochemistry Emeritus

Leona Samson
Before closing her lab, Leona Samson analyzed toxic chemicals frequently used in cancer chemotherapy to prevent further DNA damage.

Paul Schimmel
Paul Schimmel has worked throughout his career to translate bench-side research into tangible products that improve human health — including those related to alcoholism, schizophrenia, autism, AIDS, heart disease, and cancer.

Edward Scolnick
Prof Prac Emeritus
Before closing his lab, Edward Scolnick provided critical insights into the genetic underpinnings of a variety of psychiatric disorders, including bipolar disorder, schizophrenia, and autism.

Phillip A. Sharp
Before closing his lab, Phillip A. Sharp studied many aspects of gene expression in mammalian cells, including transcription, the roles of non-coding RNAs, and RNA splicing.

Ethan Signer

Before closing her lab, Hazel Sive studied fundamental mechanisms underlying vertebrate face and brain formation, as well as the molecular underpinnings for neurodevelopmental disorders.

Frank Solomon
Before closing his lab, Frank Solomon and his colleagues studied the determinants of differentiated cell morphology.

Lisa A. Steiner
Before closing her lab, Lisa A. Steiner analyzed the zebrafish genome to understand white blood cells and their role in the immune system.

JoAnne Stubbe
Before closing her lab, JoAnne Stubbe studied ribonucleotide reductases — essential enzymes that provide the building blocks for DNA replication, repair and successful targets of multiple clinical drugs.

Degree programs
Mit offers a wide range of degrees and programs..
All graduate students, whether or not they are participating in an interdepartmental program, must have a primary affiliation with and be registered in a single department. Every applicant accepted by MIT is admitted through one of the graduate departments. MIT has a number of established interdepartmental programs, and there are many more opportunities for students to arrange interdepartmental programs with interested faculty members.
All MIT graduate degree programs have residency requirements, which reflect academic terms (excluding summer). Some degrees also require completion of an acceptable thesis prepared in residence at MIT, unless special permission is granted for part of the thesis work to be accomplished elsewhere. Other degrees require a pro-seminar or capstone experience.
Applicants interested in graduate education should apply to the department or graduate program conducting research in the area of interest. Below is an alphabetical list of all the available departments and programs that offer a graduate-level degree.
Interested in reading first-hand accounts of MIT graduate students from a variety of programs? Visit the Grad Blog . Prospective students who want to talk with a current student can reach out to their department(s) of interest for connections or, if they are interested in the MIT experience for diverse communities, can reach out to a GradDiversity Ambassador .
Search Programs
This site uses cookies to give you the best possible experience. By browsing our website, you agree to our use of cookies.
If you require further information, please visit the Privacy Policy page.
- Skip to Content
- Bulletin Home

- Interdisciplinary Programs >
- Graduate Programs >
Computer Science and Molecular Biology
- Around Campus
- Academic Program
- Administration
- Arts at MIT
- Campus Media
- Fraternities, Sororities, and Independent Living Groups
- Medical Services
- Priscilla King Gray Public Service Center
- Religious Organizations
- Student Government
- Work/Life and Family Resources
- Advising and Support
- Digital Learning
- Disability and Access Services
- Information Systems and Technology
- Student Financial Services
- Writing and Communication Center
- Major Course of Study
- General Institute Requirements
- Independent Activites Period
- Undergraduate Research Opportunities Program
- First-Year Advising Seminars
- Interphase EDGE/x
- Edgerton Center
- Grading Options
- Study at Other Universities
- Internships Abroad
- Career Advising and Professional Development
- Teacher Licensure and Education
- ROTC Programs
- Financial Aid
- Medical Requirements
- Graduate Study at MIT
- General Degree Requirements
- Other Institutions
- Registration
- Term Regulations and Examination Policies
- Academic Performance and Grades
- Policies and Procedures
- Privacy of Student Records
- Abdul Latif Jameel Poverty Action Lab
- Art, Culture, and Technology Program
- Broad Institute of MIT and Harvard
- Center for Archaeological Materials
- Center for Bits and Atoms
- Center for Clinical and Translational Research
- Center for Collective Intelligence
- Center for Computational Science and Engineering
- Center for Constructive Communication
- Center for Energy and Environmental Policy Research
- Center for Environmental Health Sciences
- Center for Global Change Science
- Center for International Studies
- Center for Real Estate
- Center for Transportation & Logistics
- Computer Science and Artificial Intelligence Laboratory
- Concrete Sustainability Hub
- D-Lab
- Deshpande Center for Technological Innovation
- Division of Comparative Medicine
- Haystack Observatory
- Initiative on the Digital Economy
- Institute for Medical Engineering and Science
- Institute for Soldier Nanotechnologies
- Institute for Work and Employment Research
- Internet Policy Research Initiative
- Joint Program on the Science and Policy of Global Change
- Knight Science Journalism Program
- Koch Institute for Integrative Cancer Research
- Laboratory for Financial Engineering
- Laboratory for Information and Decision Systems
- Laboratory for Manufacturing and Productivity
- Laboratory for Nuclear Science
- Legatum Center for Development and Entrepreneurship
- Lincoln Laboratory
- Martin Trust Center for MIT Entrepreneurship
- Materials Research Laboratory
- McGovern Institute for Brain Research
- Microsystems Technology Laboratories
- MIT Center for Art, Science & Technology
- MIT Energy Initiative
- MIT Environmental Solutions Initiative
- MIT Kavli Institute for Astrophysics and Space Research
- MIT Media Lab
- MIT Office of Innovation
- MIT Open Learning
- MIT Portugal Program
- MIT Professional Education
- MIT Sea Grant College Program
- Nuclear Reactor Laboratory
- Operations Research Center
- Picower Institute for Learning and Memory
- Plasma Science and Fusion Center
- Research Laboratory of Electronics
- Simons Center for the Social Brain
- Singapore-MIT Alliance for Research and Technology Centre
- Sociotechnical Systems Research Center
- Whitehead Institute for Biomedical Research
- Women's and Gender Studies Program
- Architecture (Course 4)
- Art and Design (Course 4-B)
- Art, Culture, and Technology (SM)
- Media Arts and Sciences
- Planning (Course 11)
- Urban Science and Planning with Computer Science (Course 11-6)
- Aerospace Engineering (Course 16)
- Engineering (Course 16-ENG)
- Biological Engineering (Course 20)
- Chemical Engineering (Course 10)
- Chemical-Biological Engineering (Course 10-B)
- Chemical Engineering (Course 10-C)
- Engineering (Course 10-ENG)
- Engineering (Course 1-ENG)
- Electrical Engineering and Computer Science (Course 6-2)
- Electrical Science and Engineering (Course 6-1)
- Computation and Cognition (Course 6-9)
- Computer Science and Engineering (Course 6-3)
- Computer Science and Molecular Biology (Course 6-7)
- Electrical Engineering and Computer Science (MEng)
- Computer Science and Molecular Biology (MEng)
- Health Sciences and Technology
- Archaeology and Materials (Course 3-C)
- Materials Science and Engineering (Course 3)
- Materials Science and Engineering (Course 3-A)
- Materials Science and Engineering (PhD)
- Mechanical Engineering (Course 2)
- Mechanical and Ocean Engineering (Course 2-OE)
- Engineering (Course 2-A)
- Nuclear Science and Engineering (Course 22)
- Engineering (Course 22-ENG)
- Anthropology (Course 21A)
- Comparative Media Studies (CMS)
- Writing (Course 21W)
- Economics (Course 14-1)
- Mathematical Economics (Course 14-2)
- Data, Economics, and Design of Policy (MASc)
- Economics (PhD)
- Global Studies and Languages (Course 21G)
- History (Course 21H)
- Linguistics and Philosophy (Course 24-2)
- Philosophy (Course 24-1)
- Linguistics (SM)
- Literature (Course 21L)
- Music (Course 21M-1)
- Theater Arts (Course 21M-2)
- Political Science (Course 17)
- Science, Technology, and Society/Second Major (STS)
- Business Analytics (Course 15-2)
- Finance (Course 15-3)
- Management (Course 15-1)
- Biology (Course 7)
- Chemistry and Biology (Course 5-7)
- Brain and Cognitive Sciences (Course 9)
- Chemistry (Course 5)
- Earth, Atmospheric and Planetary Sciences (Course 12)
- Mathematics (Course 18)
- Mathematics with Computer Science (Course 18-C)
- Physics (Course 8)
- Department of Electrical Engineering and Computer Science
- Institute for Data, Systems, and Society
- Chemistry and Biology
- Climate System Science and Engineering
- Computation and Cognition
- Computer Science, Economics, and Data Science
- Humanities and Engineering
- Humanities and Science
- Urban Science and Planning with Computer Science
- African and African Diaspora Studies
- American Studies
- Ancient and Medieval Studies
- Applied International Studies
- Asian and Asian Diaspora Studies
- Biomedical Engineering
- Energy Studies
- Entrepreneurship and Innovation
- Environment and Sustainability
- Latin American and Latino/a Studies
- Middle Eastern Studies
- Polymers and Soft Matter
- Public Policy
- Russian and Eurasian Studies
- Statistics and Data Science
- Women's and Gender Studies
- Advanced Urbanism
- Computational and Systems Biology
- Computational Science and Engineering
- Design and Management (IDM & SDM)
- Joint Program with Woods Hole Oceanographic Institution
- Leaders for Global Operations
- Microbiology
- Music Technology and Computation
- Operations Research
- Real Estate Development
- Social and Engineering Systems
- Supply Chain Management
- Technology and Policy
- Transportation
- School of Architecture and Planning
- School of Engineering
- Aeronautics and Astronautics Fields (PhD)
- Artificial Intelligence and Decision Making (Course 6-4)
- Biological Engineering (PhD)
- Nuclear Science and Engineering (PhD)
- School of Humanities, Arts, and Social Sciences
- Humanities (Course 21)
- Humanities and Engineering (Course 21E)
- Humanities and Science (Course 21S)
- Sloan School of Management
- School of Science
- Brain and Cognitive Sciences (PhD)
- Earth, Atmospheric and Planetary Sciences Fields (PhD)
- Interdisciplinary Programs (SB)
- Climate System Science and Engineering (Course 1-12)
- Computer Science, Economics, and Data Science (Course 6-14)
- Interdisciplinary Programs (Graduate)
- Computation and Cognition (MEng)
- Computational Science and Engineering (SM)
- Computational Science and Engineering (PhD)
- Computer Science, Economics, and Data Science (MEng)
- Leaders for Global Operations (MBA/SM and SM)
- Music Technology and Computation (SM and MASc)
- Real Estate Development (SM)
- Statistics (PhD)
- Supply Chain Management (MEng and MASc)
- Technology and Policy (SM)
- Transportation (SM)
- Aeronautics and Astronautics (Course 16)
- Aerospace Studies (AS)
- Civil and Environmental Engineering (Course 1)
- Comparative Media Studies / Writing (CMS)
- Comparative Media Studies / Writing (Course 21W)
- Computational and Systems Biology (CSB)
- Computational Science and Engineering (CSE)
- Concourse (CC)
- Data, Systems, and Society (IDS)
- Earth, Atmospheric, and Planetary Sciences (Course 12)
- Economics (Course 14)
- Edgerton Center (EC)
- Electrical Engineering and Computer Science (Course 6)
- Engineering Management (EM)
- Experimental Study Group (ES)
- Global Languages (Course 21G)
- Health Sciences and Technology (HST)
- Linguistics and Philosophy (Course 24)
- Management (Course 15)
- Media Arts and Sciences (MAS)
- Military Science (MS)
- Music and Theater Arts (Course 21M)
- Naval Science (NS)
- Science, Technology, and Society (STS)
- Special Programs
- Supply Chain Management (SCM)
- Urban Studies and Planning (Course 11)
- Women's and Gender Studies (WGS)
Master of Engineering in Computer Science and Molecular Biology (Course 6-7P)
The Department of Biology and the Department of Electrical Engineering and Computer Science (EECS) offer a joint curriculum that focuses on the emerging field of computational and molecular biology. The curriculum provides strong foundations in both biology and computer science and features innovative, integrative, capstone, and elective subjects. The goal is to produce an entirely new cadre of graduates who are uniquely qualified to address the challenges and opportunities at the interface of computational and molecular biology. Students in the program are full members of both departments and of two schools, Science and Engineering, with one academic advisor from each department.
The Master of Engineering in Computer Science and Molecular Biology program builds on the Bachelor of Science in Computer Science and Molecular Biology program (Course 6-7) , which prepares students for careers that leverage computational biology (e.g., pharmaceuticals, bioinformatics, medicine, etc.) as well as further graduate study in biology, in computer science, and in emerging programs at the interface of these fields. The master's program provides additional depth in computational and/or molecular biology through coursework and a substantial thesis. The student selects (with departmental review and approval) 42 units of advanced graduate subjects, which include two concentration subjects in biology and/or computational biology plus a third subject in electrical engineering and computer science and/or biology. A further 24 units of electives are chosen from a restricted departmental list of math electives.
The Master of Engineering degree also requires 24 units of thesis credit. While a student may register for more than this number of thesis units, only 24 units count toward the degree requirement.
Recipients of a Master of Engineering degree normally receive a Bachelor of Science degree simultaneously. No thesis is explicitly required for the Bachelor of Science degree. However, every program must include a major project experience at an advanced level, culminating in written and oral reports. Normally, the thesis for the Master of Engineering degree will provide this experience for students receiving both degrees simultaneously.
Programs leading to the five-year Master of Engineering degree or to the four-year Bachelor of Science degree can be arranged to be identical through the junior year. At the end of the junior year, students with a strong academic record will be offered the opportunity to continue through the five-year master's program. A student in the Master of Engineering program must be registered as a graduate student for at least one regular (non-summer) term. To remain in the program and to receive the Master of Engineering degree, students will be expected to maintain a strong academic record. Admission to the Master of Engineering program is open only to undergraduate students who have completed their junior year in the Course 6-7 Bachelor of Science program.
Financial Support
The fifth year of study toward the Master of Engineering degree can be supported by a combination of personal funds, an award such as a National Science Foundation Fellowship, a fellowship, or a graduate assistantship. Assistantships require participation in research or teaching in the department or in one of the associated laboratories. Full-time assistants may register for no more than two scheduled classroom or laboratory subjects during the term, but may receive academic credit for their participation in the teaching or research program. Support through an assistantship may extend the period required to complete the Master of Engineering program by an additional term or two. Support is granted competitively to graduate students and will not be available for all of those admitted to the Master of Engineering program. If provided, department support for Master of Engineering candidates is normally limited to the first three terms as a graduate student, unless the Master of Engineering thesis has been completed or the student has served as a teaching assistant or has been admitted to the doctoral program, in which cases a fourth term of support may be permitted.
Information about these programs is available from the EECS Undergraduate Office , Room 38-476, 617-253-4654, and the Biology Undergraduate Office , Room 68-120, 617-253-4718.

Print this page.
The PDF includes all information on this page and its related tabs. Subject (course) information includes any changes approved for the current academic year.
Suggestions or feedback?
MIT News | Massachusetts Institute of Technology
- Machine learning
- Social justice
- Black holes
- Classes and programs
Departments
- Aeronautics and Astronautics
- Brain and Cognitive Sciences
- Architecture
- Political Science
- Mechanical Engineering
Centers, Labs, & Programs
- Abdul Latif Jameel Poverty Action Lab (J-PAL)
- Picower Institute for Learning and Memory
- Lincoln Laboratory
- School of Architecture + Planning
- School of Engineering
- School of Humanities, Arts, and Social Sciences
- Sloan School of Management
- School of Science
- MIT Schwarzman College of Computing
Unlocking mRNA’s cancer-fighting potential
Press contact :.

Previous image Next image
What if training your immune system to attack cancer cells was as easy as training it to fight Covid-19? Many people believe the technology behind some Covid-19 vaccines, messenger RNA, holds great promise for stimulating immune responses to cancer.
But using messenger RNA, or mRNA, to get the immune system to mount a prolonged and aggressive attack on cancer cells — while leaving healthy cells alone — has been a major challenge.
The MIT spinout Strand Therapeutics is attempting to solve that problem with an advanced class of mRNA molecules that are designed to sense what type of cells they encounter in the body and to express therapeutic proteins only once they have entered diseased cells.
“It’s about finding ways to deal with the signal-to-noise ratio, the signal being expression in the target tissue and the noise being expression in the non-target tissue,” Strand CEO Jacob Becraft PhD ’19 explains. “Our technology amplifies the signal to express more proteins for longer while at the same time effectively eliminating the mRNA’s off-target expression.”
Strand is set to begin its first clinical trial in April, which is testing a self-replicating mRNA molecule’s ability to express immune signals directly from a tumor, triggering the immune system to attack and kill the tumor cells directly. It’s also being tested as a possible improvement for existing treatments to a number of solid tumors.
As they work to commercialize its early innovations, Strand’s team is continuing to add capabilities to what it calls its “programmable medicines,” improving mRNA molecules’ ability to sense their environment and generate potent, targeted responses where they’re needed most.
“Self-replicating mRNA was the first thing that we pioneered when we were at MIT and in the first couple years at Strand,” Becraft says. “Now we’ve also moved into approaches like circular mRNAs, which allow each molecule of mRNA to express more of a protein for longer, potentially for weeks at a time. And the bigger our cell-type specific datasets become, the better we are at differentiating cell types, which makes these molecules so targeted we can have a higher level of safety at higher doses and create stronger treatments.”
Making mRNA smarter
Becraft got his first taste of MIT as an undergraduate at the University of Illinois when he secured a summer internship in the lab of MIT Institute Professor Bob Langer.
“That’s where I learned how lab research could be translated into spinout companies,” Becraft recalls.
The experience left enough of an impression on Becraft that he returned to MIT the next fall to earn his PhD, where he worked in the Synthetic Biology Center under professor of bioengineering and electrical engineering and computer science Ron Weiss. During that time, he collaborated with postdoc Tasuku Kitada to create genetic “switches” that could control protein expression in cells.
Becraft and Kitada realized their research could be the foundation of a company around 2017 and started spending time in the Martin Trust Center for MIT Entrepreneurship. They also received support from MIT Sandbox and eventually worked with the Technology Licensing Office to establish Strand’s early intellectual property.
“We started by asking, where is the highest unmet need that also allows us to prove out the thesis of this technology? And where will this approach have therapeutic relevance that is a quantum leap forward from what anyone else is doing?” Becraft says. “The first place we looked was oncology.”
People have been working on cancer immunotherapy, which turns a patient’s immune system against cancer cells, for decades. Scientists in the field have developed drugs that produce some remarkable results in patients with aggressive, late-stage cancers. But most next-generation cancer immunotherapies are based on recombinant (lab-made) proteins that are difficult to deliver to specific targets in the body and don’t remain active for long enough to consistently create a durable response.
More recently, companies like Moderna, whose founders also include MIT alumni , have pioneered the use of mRNAs to create proteins in cells. But to date, those mRNA molecules have not been able to change behavior based on the type of cells they enter, and don’t last for very long in the body.
“If you’re trying to engage the immune system with a tumor cell, the mRNA needs to be expressing from the tumor cell itself, and it needs to be expressing over a long period of time,” Becraft says. “Those challenges are hard to overcome with the first generation of mRNA technologies.”
Strand has developed what it calls the world’s first mRNA programming language that allows the company to specify the tissues its mRNAs express proteins in.
“We built a database that says, ‘Here are all of the different cells that the mRNA could be delivered to, and here are all of their microRNA signatures,’ and then we use computational tools and machine learning to differentiate the cells,” Becraft explains. “For instance, I need to make sure that the messenger RNA turns off when it's in the liver cell, and I need to make sure that it turns on when it's in a tumor cell or a T-cell.”
Strand also uses techniques like mRNA self-replication to create more durable protein expression and immune responses.
“The first versions of mRNA therapeutics, like the Covid-19 vaccines, just recapitulate how our body’s natural mRNAs work,” Becraft explains. “Natural mRNAs last for a few days, maybe less, and they express a single protein. They have no context-dependent actions. That means wherever the mRNA is delivered, it’s only going to express a molecule for a short period of time. That’s perfect for a vaccine, but it’s much more limiting when you want to create a protein that’s actually engaging in a biological process, like activating an immune response against a tumor that could take many days or weeks.”
Technology with broad potential
Strand’s first clinical trial is targeting solid tumors like melanoma and triple-negative breast cancer. The company is also actively developing mRNA therapies that could be used to treat blood cancers.
“We’ll be expanding into new areas as we continue to de-risk the translation of the science and create new technologies,” Becraft says.
Strand plans to partner with large pharmaceutical companies as well as investors to continue developing drugs. Further down the line, the founders believe future versions of its mRNA therapies could be used to treat a broad range of diseases.
“Our thesis is: amplified expression in specific, programmed target cells for long periods of time,” Becraft says. “That approach can be utilized for [immunotherapies like] CAR T-cell therapy, both in oncology and autoimmune conditions. There are also many diseases that require cell-type specific delivery and expression of proteins in treatment, everything from kidney disease to types of liver disease. We can envision our technology being used for all of that.”
Share this news article on:
Related links.
- Strand Therapeutics
- Department of Biological Engineering
Related Topics
- Biological engineering
- Bioengineering and biotechnology
- Drug development
- Tissue engineering
- Synthetic biology
- Innovation and Entrepreneurship (I&E)
Related Articles
Synthetic biology circuits can respond within seconds
MIT launches Center for Multi-Cellular Engineered Living Systems
This RNA-based technique could make gene therapy more effective
Previous item Next item
More MIT News

Most work is new work, long-term study of U.S. census data shows
Read full story →
Does technology help or hurt employment?
A first-ever complete map for elastic strain engineering

Shining a light on oil fields to make them more sustainable

“Life is short, so aim high”

MIT launches Working Group on Generative AI and the Work of the Future
- More news on MIT News homepage →
Massachusetts Institute of Technology 77 Massachusetts Avenue, Cambridge, MA, USA
- Map (opens in new window)
- Events (opens in new window)
- People (opens in new window)
- Careers (opens in new window)
- Accessibility
- Social Media Hub
- MIT on Facebook
- MIT on YouTube
- MIT on Instagram

College of Science
Bioinformatics.
Northeastern University’s Master of Science program in Bioinformatics provides cross-disciplinary training in biology, computer science, and informational technology for today’s cutting-edge jobs in the biotechnology and pharmaceutical industries.
Northeastern University is committed to delivering cutting-edge programs that foster interdisciplinary thinking, research, and the pursuit of innovation-driven discoveries that have an impact on lives. That commitment and vision are at the very core of the MS in Bioinformatics. The program’s cross-disciplinary training prepares graduates to succeed in multiple roles in this new and growing field.
Combining challenging academics in biology, computer science, and information technology with real-world experience, the program helps students integrate the knowledge, skills, experience, and confidence they need to achieve their goals and make a difference in our world. The Master of Science in Bioinformatics is structured to provide students with the skills and knowledge to develop, evaluate, and deploy bioinformatics and computational biology applications. The program is designed to prepare students for employment in the biotechnology sector, where the need for knowledgeable life scientists with quantitative and computational skills has exploded in the past decade.
Concentrations:
- Bioinformatics Enterprise: The Bioinformatics Enterprise concentration integrates business and management skills with the science of bioinformatics. Students learn the fundamental concepts of leadership, entrepreneurship and innovation, financial decision making, and marketing. They gain teamwork, management, and business development skills in the process and graduate prepared to become scientist-managers.
- Biotechnology: The Biotechnology concentration provides students without a biotechnology background to obtain a strong foundation in basic biotechnology concepts and skills. Individuals, particularly those who are working in fields other than biotechnology, will acquire competency and learn new practical skills enabling them to increase productivity and allow for transitions into more biotechnology-related fields.
- Data Analytics: The Data Analytics concentration is designed to provide students with foundational knowledge in data science—including data management, machine learning, data mining, statistics, and visualizing and communicating data—that can be applied to data-driven decision making in any discipline.
- Health Informatics: The Health Informatics concentration will help prepare students to successfully address the combined clinical, technical, and business needs of health-related professionals.
- Medical Health Informatics: The Medical Health Informatics concentration will help prepare students to successfully address the combined medical needs from a patient's health perspective, technical, and business needs.
- Omics: The omics concentration will prepare students to analyze large data sets related to genomics, proteomics, transcriptomics and new-omics fields as they evolve.
- Coursework Option: Students are not required to declare a concentration. With the elective option, students select 12 credits of electives in place of concentration-specific courses.
More Details
Unique features.
- Our online format allows students to participate in the program anywhere in the world
- Students gain up to six months of work experience through co-op position
- With only one additional class, a student can also earn a graduate certificate in data science
- 94% employment after graduation in industry or research in last three years
Program Objectives
- Attain core knowledge in bioinformatics programming
- Integrate knowledge from biological, computational, and mathematical disciplines
- Gain professional work experience via co-op
Looking for something different?
A graduate degree or certificate from Northeastern—a top-ranked university—can accelerate your career through rigorous academic coursework and hands-on professional experience in the area of your interest. Apply now—and take your career to the next level.
Program Costs
Finance Your Education We offer a variety of resources, including scholarships and assistantships.
How to Apply Learn more about the application process and requirements.
Requirements
- Online application
- Application fee
- Transcripts from all institutions attended
- Personal statement
- 2 letters of recommendation
- GRE not required
- Degree earned or in progress at a U.S / Canadian institution
- Degree earned or in progress at an institution where English is the only medium of instruction
- Official exam scores from either the TOEFL iBT (institution code is 3682), IELTS, PTE exam, or Duolingo English Test. Scores are valid for 2 years from the test date.
Learn more about applying to the College of Science.
Are You an International Student? Find out what additional documents are required to apply.
Admissions Details Learn more about the College of Science admissions process, policies, and required materials.
Admissions Dates
Learn more about applying to the College of Science and our admissions deadlines.
Industry-aligned courses for in-demand careers.
For 100+ years, we’ve designed our programs with one thing in mind—your success. Explore the current program requirements and course descriptions, all designed to meet today’s industry needs and must-have skills.
View curriculum
Students in the MS in Bioinformatics program gain real-world knowledge, awareness, perspective, and confidence during a three- or six-month graduate co-op in industry or academia. As a recognized leader in experiential learning and a trusted source of high-caliber students, Northeastern enjoys relationships with more than 3,000 public- and private-sector employers on seven continents. Recent bioinformatics co-op partners have included:
- Broad Institute
- Harvard Medical School
- Brigham and Women’s Hospital
- Dana-Farber Cancer Institute
- Seattle Children’s Research Institute
Our Faculty
Northeastern University faculty represents a broad cross-section of professional practices and fields, including finance, education, biomedical science, management, and the U.S. military. They serve as mentors and advisors and collaborate alongside you to solve the most pressing global challenges facing established and emerging markets.

By enrolling in Northeastern, you’ll gain access to students at 13 campus locations, 300,000+ alumni, and 3,000 employer partners worldwide. Our global university system provides students unique opportunities to think locally and act globally while serving as a platform for scaling ideas, talent, and solutions.
Below is a look at where our Science & Mathematics alumni work, the positions they hold, and the skills they bring to their organization.
Where They Work
- State Street
- Liberty Mutual Insurance
What They Do
- Engineering
- Business Development
- Information Technology
What They're Skilled At
- Project Management
- Data Analysis
Learn more about Northeastern Alumni on Linkedin.
Related Articles

Computational Biology vs. Bioinformatics: What’s the Difference?

Earning a Bioinformatics Degree: Career Outlook and Job Prospects

7 Popular Bioinformatics Careers
- Postdoc India
- Postdoc Abroad
- Postdoc (SS)
- RESEARCHERSJOB
- Post a position
- JRF/SRF/Project
- Science News

International PhD Positions: Computational Biology, Leibniz Institute, Germany

PhD Positions: Computational Biology: Join our dynamic team in Computational Biology! We are currently seeking motivated candidates for various positions, including a bioinformatician role and international PhD openings. Additionally, if you have your own funding or are interested in applying for fellowships, we welcome collaborations in developing hybrid projects. Explore the intersection of aging, microbiome, and computational biology with us and contribute to cutting-edge research in an innovative environment.
Study Area: Computational Biology, Bioinformatics, Computer Science, Biology, Wetlab-Drylab Hybrid Projects
Location: Leibniz Institute, Germany
Eligibility/Qualification: For the bioinformatician position, candidates should hold a PhD in computational biology, bioinformatics, computer science, biology, or related fields. Exceptional candidates without a PhD but with significant hands-on experience will also be considered. Proficiency in R, Python, Bash, and version control is required, along with experience in high-throughput data preprocessing and analysis. Excellent communication skills in English are essential.
For international PhD positions, interested candidates should refer to the DAAD-supported openings and submit applications according to the institute’s guidelines. Candidates with backgrounds in computational biology or mixed wet-dry backgrounds are encouraged to apply. PhD and postdoc candidates with their own funding or interest in applying for fellowships are welcome to explore opportunities for collaboration.
Description: As a bioinformatician in our team, you will be responsible for establishing and running computational pipelines for preprocessing and integrating datasets from various high-throughput experiments. You will collaborate with other computational biologists and experimentalists to support research data management and collaborative projects. For international PhD applicants, positions are available with DAAD support across multiple groups, including ours. We encourage applicants with computational biology backgrounds or mixed wet-dry backgrounds to apply and list our group as a preferred choice.
If you have your own funding or are interested in applying for fellowships, we invite you to collaborate with us on developing hybrid projects at the intersection of aging, microbiome, and computational biology. Prestigious fellowship opportunities such as MSCA Postdoctoral Fellowships, EMBO Postdoctoral Fellowships, and HFSP LTF are available, requiring high knowledge transfer. We welcome postdoctoral candidates from non-biological disciplines as well.
How to Apply: Interested candidates for the bioinformatician position should check the ad on our institute website and submit their applications according to the provided instructions. For international PhD positions, applicants should refer to the institute’s website for more information and submit applications accordingly. Candidates with their own funding or interested in fellowships should contact Melike directly with their CV, cover letter, references, Google Scholar link, and a brief project description.
- For the bioinformatician position, the application deadline is May 5th, 2024.
- For international PhD positions, the application deadline is May 12th, 2024.
- For fellowship opportunities, please refer to specific fellowship deadlines mentioned in the description.
RELATED ARTICLES MORE FROM AUTHOR
Postdoctoral researcher and phd positions, forschungszentrum jülich, germany, fully funded ph.d. position: neurodiverse individuals, campus lille, france, phd scholarship: asymmetric catalysis, institut parisien de chimie moléculaire, paris, france, phd opportunity: inorganic nanomaterials at okayama university, japan, phd position – complex biogenesis, heidelberg university, germany, phd position in amo physics -vrije universiteit amsterdam, netherlands, leave a reply cancel reply.
Save my name, email, and website in this browser for the next time I comment.
Follow us on Instagram @researchersjob_rj
- Terms Of Service
- Privacy Policy
Postdoctoral Associate in Molecular Biophysics, University of Oxford, UK

- Who’s Teaching What
- Subject Updates
- MEng program
- Opportunities
- Minor in Computer Science
- Resources for Current Students
- Program objectives and accreditation
- Graduate program requirements
- Admission process
- Degree programs
- Graduate research
- EECS Graduate Funding
- Resources for current students
- Student profiles
- Instructors
- DEI data and documents
- Recruitment and outreach
- Community and resources
- Get involved / self-education
- Rising Stars in EECS
- Graduate Application Assistance Program (GAAP)
- MIT Summer Research Program (MSRP)
- Sloan-MIT University Center for Exemplary Mentoring (UCEM)
- Electrical Engineering
- Computer Science
- Artificial Intelligence + Decision-making
- AI and Society
- AI for Healthcare and Life Sciences
- Artificial Intelligence and Machine Learning
- Biological and Medical Devices and Systems
- Communications Systems
- Computational Biology
- Computational Fabrication and Manufacturing
- Computer Architecture
- Educational Technology
- Electronic, Magnetic, Optical and Quantum Materials and Devices
- Graphics and Vision
- Human-Computer Interaction
- Information Science and Systems
- Integrated Circuits and Systems
- Nanoscale Materials, Devices, and Systems
- Natural Language and Speech Processing
- Optics + Photonics
- Optimization and Game Theory
- Programming Languages and Software Engineering
- Quantum Computing, Communication, and Sensing
- Security and Cryptography
- Signal Processing
- Systems and Networking
- Systems Theory, Control, and Autonomy
- Theory of Computation
- Departmental History
- Departmental Organization
- Visiting Committee
- News & Events
Global Disability Innovation Hub
- News & Events
- EECS Celebrates Awards

New software enables blind and low-vision users to create interactive, accessible charts
Screen-reader users can upload a dataset and create customized data representations that combine visualization, textual description, and sonification.
- DACA/Undocumented
- First Generation, Low Income
- International Students
- Students of Color
- Students with disabilities
- Undergraduate Students
- Master’s Students
- PhD Students
- Faculty/Staff
- Family/Supporters
- Career Fairs
- Post Jobs, Internships, Fellowships
- Build your Brand at MIT
- Recruiting Guidelines and Resources
- Connect with Us
- Career Advising
- Distinguished Fellowships
- Employer Relations
- Graduate Student Professional Development
- Prehealth Advising
- Student Leadership Opportunities
- Academia & Education
- Architecture, Planning, & Design
- Arts, Communications, & Media
- Business, Finance, & Fintech
- Computing & Computer Technology
- Data Science
- Energy, Environment, & Sustainability
- Life Sciences, Biotech, & Pharma
- Manufacturing & Transportation
- Health & Medical Professions
- Social Impact, Policy, & Law
- Getting Started & Handshake 101
- Exploring careers
- Networking & Informational Interviews
- Connecting with employers
- Resumes, cover letters, portfolios, & CVs
- Finding a Job or Internship
- Post-Graduate and Summer Outcomes
- Professional Development Competencies
- Preparing for Graduate & Professional Schools
- Preparing for Medical / Health Profession Schools
- Interviewing
- New jobs & career transitions
- Career Prep and Development Programs
- Employer Events
- Outside Events for Career and Professional Development
- Events Calendar
- Career Services Workshop Requests
- Early Career Advisory Board
- Peer Career Advisors
- Student Staff
- Mission, Vision, Values and Diversity Commitments
- News and Reports
Envisagenics, Inc.
Computational biology intern.
- Share This: Share Computational Biology Intern on Facebook Share Computational Biology Intern on LinkedIn Share Computational Biology Intern on X
Job Description: Computational Biology Intern at Envisagenics
Position: Computational Biology Intern
Company: Envisagenics
Location: New York – remote work available
Employment Type: part time internship. 10 weeks
About Envisagenics:
Envisagenics, Inc. is an RNA therapeutics company located in New York City. The company uses Artificial Intelligence to help find cures to diseases caused by RNA splicing errors. The mission of Envisagenics is to materially advance the world’s ability to cure diseases by discovering new and more efficient drug targets through the analysis and integration of genomic data alongside our pharma partners.
Job Summary:
We are seeking a skilled and motivated Computational Biology Intern. As a Computational Biology Intern at Envisagenics, you will contribute to the development and application of computational methodologies to analyze large-scale RNA sequencing datasets. You will collaborate closely with a growing team of computational biologists, data scientists, software engineers, and biologists to extract meaningful insights from complex biological data, helping us pave the way for the development of next-generation RNA-based therapeutics. We are looking for someone with experience in RNA-seq data, genomics, RNA splicing, machine learning, and/or biological data interpretation. Candidates will be responsible for developing and implementing computational approaches and performing hypothesis-driven data interrogation to make fundamental contributions to our understanding of RNA-sequencing data and disease biology and further the development of RNA-based therapeutics.
Responsibilities:
1. Develop computational algorithms and pipelines for RNA-sequencing analysis and biological interpretation.
2. Utilize statistical methods and machine learning techniques to analyze and interpret complex biological data.
3. Collaborate with experimental biologists to design experiments and provide computational support for data analysis and interpretation.
4. Stay up-to-date with the latest advancements in computational biology, bioinformatics, and RNA splicing research.
5. Contribute to the enhancement and maintenance of internal computational pipelines and databases.
6. Prepare clear and concise reports and presentations to communicate findings to the team and stakeholders.
Qualifications:
1. Current PhD student or recent PhD graduate in Computational Biology, Bioinformatics, or a related field, preferably with a focus on RNA splicing or RNA biology.
2. Extensive experience working with RNA-sequencing data.
3. Strong proficiency in programming languages such as Python and R.
4. Familiarity and comfort high-performance computing (HPC) capabilities.
5. Experience with building/supporting/using bioinformatics pipelines.
6. Experience extracting biological insights from RNA-sequencing data and other types of genomic and transcriptomic data.
7. Experience with statistical and mathematical modeling with genomic and transcriptomic data.
8. Familiarity with public genomic databases, resources, and relevant computational tools.
9. Ability to work in an agile environment.
10. Must be independent and strategically minded, and team-oriented.
11. Must have proficient written, communication and presentation skills.
12. Must be willing to show proof of vaccination.
Preferred Qualifications:
1. Experience with Microsoft Azure (or Amazon AWS).
2. Experience with SQL and MySQL.
3. Experience with collaborative software development (e.g. git)
4. Experience with C++, perl, or other scripting languages
5. Familiarity with genome browser tools and public bioinformatics databases – UCSC, IGV, NCBI, ENCODE, cBioPortal, etc.
To apply, please submit your CV and a cover letter detailing your relevant experience and interest in computational biology to [email protected]
Pay: $25 an hour. Interns are not eligible for benefits.
Envisagenics is an equal opportunity employer and welcomes applicants from diverse backgrounds.
- Management Team
- Working Groups
- Leadership Partners & Associate Members
- Pre-Clinical Courses
- Physician Associate Program
- Physician Assistant Online Program
- Yale School of Nursing
- Yale School of Public Health
- Curriculum Development
- Faculty & Staff
- Students & Trainees
- Defending Gender-Affirming Care in Alabama
- Flawed Medicaid Report in Florida
- Biased Science in Texas & Alabama
- Yale LGBTQI+ Research Resources
- Funding Options
- LGBTQI Healthcare Publications
- Education & Training Team
- Affiliate Members
- Yale Clinics & Programs
- Education & Training Inventory
- Organizations with LGBTQI Health Resources
- Our Members
- FAQs on Pronoun use
- LGBTQI+ and Diversity Groups at Yale
- Health Professional Community
- Connecticut State Community
- Regional and National Community
- News & Events
INFORMATION FOR
- Residents & Fellows
- Researchers
Steven Reilly, PhD
Contact information, lab location.
- TAC S340 The Anlyan Center 300 Cedar Street New Haven, CT 06519
Mailing Address
Yale School of Medicine
PO Box 208005
New Haven, CT 06520-8005
United States
Research & Publications
Appointments.
Steven Reilly received his B.S. in Biology from Carnegie Mellon University in 2009. Motivated by the rapid emergence of new technologies to map the full epigenomes, he joined Jim Noonan's Lab in the Genetics Department of Yale School of Medicine. There he built gene regulatory maps of the developing human, rhesus, and mouse cortex to identify changes underlying unique aspects of human brain morphology and cognitive abilities. Steve received his Ph.D. in 2015 and then joined the laboratory of Pardis Sabeti at the Broad Institute of Harvard and MIT to interrogate the function of genetic variants at the intersection of natural selection and human disease. As evolutionary adaptive genetic variants have been shown to underlie diversity in disease risk and morphology across human populations, the lens of evolution remains a powerful, yet underutilized method for understanding human biology He is specifically interested in furthering our understanding of non-coding variation, the main cache of human genetic diversity. The has created novel machine-learning methods to predict the subset of human variants under selection that are functional, and experimental methods to characterize variants in a massively parallel fashion. Steve has developed endogenous CRISPR perturbation methods and synthetic DNA technologies coupled with genomic readouts to directly assess the cellular phenotypes of non-coding alleles. Steve joined the Yale Department of Genetics as an Assistant Professor in September, 2021.
The Reilly lab develops and applies new high-throughput experimental approaches to interrogate the genome, such as non-coding CRISPR screens and the Massively Parallel Reporter Assay. Computationally, we also develop machine-learning approaches to predict the functions of these CRE perturbations. Together with these new tools, we use evolution as a powerful lens for characterizing genomic signals of positive selection that impact modern human phenotypes and diseases.
The lab has three main foci:
- Developing new, large-scale experimental screens to perturb CREs, and new computational tools to model their function
- Identifying evolutionary adaptive alleles likely impacting modern human phenotypes
- Applying these functional genomic tools to phenotypically interesting loci important for human disease and evolution.
Education & Training
- Postdoctoral Fellow Broad Institute of Harvard and MIT (2021)
- PhD Yale, Genetics (2015)
- BS Carnegie Mellon, Biology (2009)
Honors & Recognition
Departments & organizations.
- Center for RNA Science and Medicine
- Computational Biology and Biomedical Informatics
- Janeway Society
- Molecular Cell Biology, Genetics and Development
- Yale Center for Genomic Health
- Yale Combined Program in the Biological and Biomedical Sciences (BBS)

COMMENTS
The program includes teaching experience during one semester of the second year. It prepares students with the tools needed to succeed in a variety of academic and non-academic careers. The program is highly selective with typical class sizes 8 to 10 students. About half of our graduate students are women, about one-quarter are international ...
The CSB PhD program is an Institute-wide program that has been jointly developed by the Departments of Biology, Biological Engineering, and Electrical Engineering and Computer Science. The program integrates biology, engineering, and computation to address complex problems in biological systems, and CSB PhD students have the opportunity to work ...
MIT Computational and Systems Biology Graduate Admissions Statement. March 26, 2020. In response to the challenges of teaching, learning, and assessing academic performance during the global COVID-19 pandemic, MIT has adopted the following principle: MIT's admissions committees and offices for graduate and professional schools will take the significant disruptions of the COVID-19 outbreak in ...
Computational and systems biology, as practiced at MIT, is organized around "the 3 Ds" of description, distillation, and design. In many research programs, systematic data collection is used to create detailed molecular- or cellular-level descriptions of a system in one or more de ned states. Given the complexity of biological systems and the ...
Computational and Systems Biology. 77 Massachusetts Avenue. Building 68-230A. Cambridge MA, 02139. 617-324-4144. [email protected]. Website: Computational and Systems Biology. Apply here.
CSB students may choose any MIT faculty member as their PhD advisor, including those at MIT-affiliated institutes such as the Broad, Koch, Ragon, or Whitehead Institute. Massachusetts Institute of Technology. MIT Computational & Systems Biology PhD Program (CSB) Massachusetts Institute of Technology 77 Massachusetts Avenue Bldg 68, Room 120C ...
MIT Computational & Systems Biology PhD Program (CSB) Massachusetts Institute of Technology 77 Massachusetts Avenue Bldg 68, Room 120C Cambridge, MA 02139. [email protected] 617.324.4144.
The Computational and Systems Biology (CSB) Ph.D. Program prepares students to become independent, interdisciplinary researchers in post-genomic biology and related fields. There is a strong focus on quantitative methods and modeling, experimental design, and device development.
CSB.100 [J] Topics in Computational and Systems Biology. Seminar based on research literature. Papers covered are selected to illustrate important problems and varied approaches in the field of computational and systems biology, and to provide students a framework from which to evaluate new developments. Preference to first-year CSB PhD students.
CSB: Computational Systems Biology PhD Program. 617-324-4144. [email protected]
It is the only subject in the program with such a limitation. CSB.100 Topics in Computational and Systems Biology Modern Biology (One Subject): A semester of modern graduate-level biology at MIT strengthens the biology base of all students in the program. Subjects in molecular biology, neurobiology, biochemistry, or genetics fulfill this ...
Ph.D. applicants: If you are interested in applying to MIT, please consider the Computer Science (EECS) and Computational and Systems Biology (CSBi) and Biological Engineering and Health Science and Technology graduate programs. Admitted graduate students are invited for on-campus visits to MIT, which is a great opportunity to meet with faculty ...
The Ph.D. program in Computational Biology draws on course offerings from the disciplines of the Center's Core faculty members. These areas are Applied Mathematics, Computer Science, the Division of Biology and Medicine, the Center for Biomedical Informatics, and the School of Public Health. Our faculty and Director of Graduate Studies work ...
Our world-renowned faculty include 3 Nobel laureates; 29 members of the National Academy of Sciences; 11 Howard Hughes Medical Institute (HHMI) investigators; and 4 recipients of the National Medal of Science. Filter by.
All MIT graduate degree programs have residency requirements, which reflect academic terms (excluding summer). ... Computational and Systems Biology: December 1: Computational Science and Engineering PhD: December 1: Computational Science and Engineering SM: Data, Economics, and Design of Policy: January 13:
The master's program provides additional depth in computational and/or molecular biology through coursework and a substantial thesis. The student selects (with departmental review and approval) 42 units of advanced graduate subjects, which include two concentration subjects in biology and/or computational biology plus a third subject in ...
The experience left enough of an impression on Becraft that he returned to MIT the next fall to earn his PhD, where he worked in the Synthetic Biology Center under professor of bioengineering and electrical engineering and computer science Ron Weiss. ... and here are all of their microRNA signatures,' and then we use computational tools and ...
The Master of Science in Bioinformatics is structured to provide students with the skills and knowledge to develop, evaluate, and deploy bioinformatics and computational biology applications. The program is designed to prepare students for employment in the biotechnology sector, where the need for knowledgeable life scientists with quantitative ...
PhD Positions: Computational Biology: Join our dynamic team in Computational Biology! We are currently seeking motivated candidates ... phd program scholarships; PhD Programme; phd programmes; PhD Scholarship; phd scholarships; PhD Student Position; Research job; Share. Facebook. ... MIT, Cambridge, MA, USA. April 1, 2024.
Electrical Engineers design systems that sense, process, and transmit energy and information. We leverage computational, theoretical, and experimental tools to develop groundbreaking sensors and energy transducers, new physical substrates for computation, and the systems that address the shared challenges facing humanity.
Job Description: Computational Biology Intern at Envisagenics Position: Computational Biology Intern Company: Envisagenics Location: New York - remote work available Employment Type: part time …
Steven Reilly received his B.S. in Biology from Carnegie Mellon University in 2009. ... Steve received his Ph.D. in 2015 and then joined the laboratory of Pardis Sabeti at the Broad Institute of Harvard and MIT to interrogate the function of genetic variants at the intersection of natural selection and human disease. ... and new computational ...