5 Reasons to Pursue Your PhD at EMBL
When it comes to starting a successful career in life sciences, the European Molecular Biology Laboratory (EMBL) stands out as a great choice. The fully-funded EMBL International PhD Programme (EIPP) is designed to promote interdisciplinary research, international collaboration and early independence. By completing your PhD education at one of EMBL's six sites (in Barcelona, Grenoble, Hamburg, Heidelberg, Hinxton and Rome), you will develop the skills needed to excel in your future career in academia, industry, and beyond. Find out why EMBL could be the right place for you!

Engage in Interdisciplinary Research beyond State-of-the-Art
As a PhD student at EMBL, you'll have access to state-of-the-art laboratories, pioneering research and technologies, and an impressive array of multidisciplinary expertise. The organisation is dedicated to promoting excellence in molecular life sciences and advocates an interdisciplinary approach to tackle the increasingly complex biological research questions of our time. The EIPP welcomes candidates with backgrounds in biology, chemistry, physics, mathematics, molecular medicine, computer science, and other relevant disciplines to apply to work on research projects at the forefront of scientific progress.
Dive into a Truly International Environment
Known as Europe's life sciences laboratory, EMBL nurtures scientific collaboration across Europe and beyond. Ranging from the seaside views of Barcelona to the Alpine landscapes in Grenoble, the unique identities of EMBL's six sites contribute to the diversity and richness of the overall organisation while still maintaining a collaborative and cohesive environment for scientists across all locations.
At EMBL, you'll join an international PhD program where students come from around 50 different countries. This multicultural character not only fosters the exchange of innovative ideas but also offers you an opportunity to learn from people of many cultures and backgrounds.
Convinced yet?
Experience a collaborative and supportive work culture.
EMBL's strength lies in its unique approach towards science and a strong sense of community, where students and principal investigators work together to tackle challenging research questions. Extensive collaboration between the research groups across EMBL is encouraged from early on, with the current faculty and student body rating the collaborative environment as one of the top reasons for choosing EMBL.
EMBL believes that diversity is a driving factor in scientific excellence and innovation and is committed to promoting a fair, diverse and inclusive workplace, where an individual's unique background and perspective are valued. Current PhD students highlight the supportive environment that empowers individuals to thrive. As Saul Pierotti, a predoctoral fellow from EMBL-EBI puts it: "If you want to be in a scientifically stimulating environment that also values your needs as a person, I think EMBL is a great place."
Advance Your Career with EMBL's Training and Support
As an EIPP student, you will benefit from tailored training and individual career support, helping you develop the scientific and complementary skills you need for your PhD project and future career. EMBL's comprehensive training portfolio balances theory and practice, close mentoring and creative freedom, collaborative teamwork and independence. Training often comes directly from in-house experts, and programs are constantly evolving to meet the changing needs of PhD students.
Advisors from the EMBL Fellows' Career Service are available to support your career planning. The service offers information and guidance through tailored resources, group workshops and individual support. And with links to industry partners, as well as events like the annual Career Day, students have the chance to learn more about potential career options within and beyond academia.
Explore additional opportunities and build a strong network
At EMBL, the network you will build extends far beyond scientific collaborations. As a PhD student, you can engage in a wide range of activities, from attending courses and conferences to participating in outreach initiatives. There are additional opportunities to foster leadership and communication skills, including the organisation of flagship events, such as the annual PhD Symposium, becoming a student representative, or getting involved in committees and teaching activities. Through these experiences, you will forge invaluable connections with scientific peers, mentors, and professionals across different sectors, creating the right setting for you to grow and build your network.
If you're looking for an international and interdisciplinary PhD program in the life sciences, EMBL is the perfect place for you! Besides access to research and technologies beyond state-of-the-art and a truly collaborative environment, you'll benefit from tailored training and career development support, and plenty of opportunities to build a network.
Jobs by field
- Electrical Engineering 197
- Machine Learning 195
- Artificial Intelligence 190
- Programming Languages 157
- Molecular Biology 144
- Cell Biology 109
- Materials Engineering 108
- Management 107
- Electronics 107
- Engineering Physics 106
Jobs by type
- Postdoc 366
- Assistant / Associate Professor 248
- Professor 156
- Researcher 124
- Research assistant 105
- Lecturer / Senior Lecturer 95
- Tenure Track 87
- Engineer 71
- Management / Leadership 71
Jobs by country
- Belgium 293
- Netherlands 234
- Finland 130
- Switzerland 126
- Germany 118
- Luxembourg 102
Jobs by employer
- University of Luxembourg 100
- KU Leuven 89
- Eindhoven University of Techn... 86
- Mohammed VI Polytechnic Unive... 84
- Ghent University 69
- ETH Zürich 61
- University of Twente 59
- KTH Royal Institute of Techno... 51
- Wenzhou-Kean University 35
This website uses cookies

Study at Cambridge
About the university, research at cambridge.
- Undergraduate courses
- Events and open days
- Fees and finance
- Postgraduate courses
- How to apply
- Postgraduate events
- Fees and funding
- International students
- Continuing education
- Executive and professional education
- Courses in education
- How the University and Colleges work
- Term dates and calendars
- Visiting the University
- Annual reports
- Equality and diversity
- A global university
- Public engagement
- Give to Cambridge
- For Cambridge students
- For our researchers
- Business and enterprise
- Colleges & departments
- Email & phone search
- Museums & collections
- Course Directory
- Department directory
European Bioinformatics Institute
Postgraduate Study
- Why Cambridge overview
- Chat with our students
- Cambridge explained overview
- The supervision system
- Student life overview
- In and around Cambridge
- Leisure activities
- Student unions
- Music awards
- Student support overview
- Mental health and wellbeing
- Disabled students
- Accommodation
- Language tuition
- Skills training
- Support for refugees
- Courses overview
- Qualification types
- Funded studentships
- Part-time study
- Research degrees
- Visiting students
- Finance overview
- Fees overview
- What is my fee status?
- Part-time fees
- Application fee
- Living costs
- Funding overview
- Funding search
- How to apply for funding
- University funding overview
- Research Councils (UKRI)
- External funding and loans overview
- Funding searches
- External scholarships
- Charities and the voluntary sector
- Funding for disabled students
- Widening participation in funding
- Colleges overview
- What is a College?
- Choosing a College
- Terms of Residence
- Applying overview
- Before you apply
- Entry requirements
- Application deadlines
- How do I apply? overview
- Application fee overview
- Application fee waiver
- Life Science courses
- Terms and conditions
- Continuing students
- Disabled applicants
- Supporting documents overview
- Academic documents
- Finance documents
- Evidence of competence in English
- Terms and Conditions
- Applicant portal and self-service
- After you apply overview
- Confirmation of admission
- Student registry
- Previous criminal convictions
- Deferring an application
- Updating your personal details
- Appeals and Complaints
- Widening participation
- Postgraduate admissions fraud
- International overview
- Immigration overview
- ATAS overview
- Applying for an ATAS certificate
- Current Cambridge students
- International qualifications
- Competence in English overview
- What tests are accepted?
- International events
- International student views overview
- Akhila’s story
- Alex’s story
- Huijie’s story
- Kelsey’s story
- Nilesh’s story
- Get in touch!
- Events overview
- Upcoming events
- Postgraduate Open Days overview
- Discover Cambridge: Master’s and PhD Study webinars
- Virtual tour
- Research Internships
- How we use participant data
- Postgraduate Newsletter
1 course offered in the European Bioinformatics Institute
Biological science (ebi) - phd.
Established in 1983, the EMBL International PhD Programme provides students with the best starting platform for a successful career in science. Characterised by first-class training, internationality, dedicated mentoring and early independence in research, it is among the world's most competitive PhD training schemes in molecular biology. All of EMBL's six outstations participate in the programme.
EMBL-EBI provides a highly collaborative, interdisciplinary environment in which research and service provision are closely allied. We are a world leader in bioinformatics research and service provision, as we are at the centre of global efforts to collect and disseminate biological data. We share a campus with the Wellcome Sanger Institute, 12 miles south of Cambridge in the United Kingdom. EMBL PhD students at EMBL-EBI are members of the University of Cambridge and one of its Colleges. They receive their degree from Cambridge University; the programme is coordinated in Heidelberg with local support at EBI. Please visit the EMBL International PhD Programme pages to learn about how to apply. Please note all applicants must secure a place on the EMBL programme before submitting an application to the University of Cambridge.
More Information
1 course also advertised in the European Bioinformatics Institute
Clinical medicine wellcome trust - phd - closed.
From the Faculty of Clinical Medicine
We provide high-quality research training to clinical health professionals with an aptitude for research to enable them to become future leaders in medical and healthcare science. We offer training in an outstanding environment, spanning basic science, translational medicine, interdisciplinary, behavioural and applied health research.
We take great pride in our track record of successfully training health professionals to undertake the highest quality research across Cambridge and Norwich. We offer one of the most rewarding environments in which you could pursue your research training with world-leading researchers in The Schools of Clinical Medicine and Biological Sciences at the Universities of Cambridge, Wellcome Sanger Institute and other MRC, Wellcome & Cancer Research UK funded Institutes, Centres & Units in the wider Cambridge area, as well as the School of Health Sciences and Norwich Medical School at the University of East Anglia with other partners on the Norwich Research Park. The most important criteria we are looking for are the pursuit of research excellence, hard work and the will to make a difference to health.
The programme faculty provides mentoring and guidance on opportunities to undertake pre-doctoral research placements, enabling successful candidates to make an informed choice of PhD project and supervisor. Bespoke training and support for career development for fellows, together with support to supervisors, ensures a successful research experience. Post-doctorally, we will guide fellows based on their individual progress, to make the transition into higher research fellowships and clinical pathways, enabling ongoing training with continuance of research momentum.
Department Members
Dr rolf apweiler and dr ewan birney director.
- 16 Academic Staff
- 40 Postdoctoral Researchers
- 43 Graduate Students
http://www.ebi.ac.uk/
Postgraduate admissions office.
- Admissions Statistics
- Start an Application
- Applicant Self-Service
At a glance
- Bringing a family
- Current Postgraduates
- Cambridge Students' Union (SU)
University Policy and Guidelines
Privacy Policy
Information compliance
Equality and Diversity
Terms of Study
About this site
About our website
Privacy policy
© 2024 University of Cambridge
- Contact the University
- Accessibility
- Freedom of information
- Privacy policy and cookies
- Statement on Modern Slavery
- University A-Z
- Undergraduate
- Postgraduate
- Research news
- About research at Cambridge
- Spotlight on...
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- PMC10666930

The changing career paths of PhDs and postdocs trained at EMBL
1 Genome Biology Unit, European Molecular Biology Laboratory Heidelberg, Germany
Britta Velten
Bernd klaus, mauricio ramm.
2 EMBL International Centre for Advanced Training, European Molecular Biology Laboratory Heidelberg, Germany
Wolfgang Huber
Rachel coulthard-graf, associated data.
The data were collated for the provision of statistics, and are stored in a manner compliant with EMBL's internal policy on data protection . This policy means that the full dataset cannot be made publicly available (because the nature of the data means that sufficient anonymisation is not possible). Summary statistics for the main data table can be found in Supplementary file 1 (Table S1). Rmarkdown documentation of the analysis and figures can be found here and is available on GitHub (copy archived at Coulthard and Lu, 2022 ).
Individuals with PhDs and postdoctoral experience in the life sciences can pursue a variety of career paths. Many PhD students and postdocs aspire to a permanent research position at a university or research institute, but competition for such positions has increased. Here, we report a time-resolved analysis of the career paths of 2284 researchers who completed a PhD or a postdoc at the European Molecular Biology Laboratory (EMBL) between 1997 and 2020. The most prevalent career outcome was Academia: Principal Investigator (636/2284=27.8% of alumni), followed by Academia: Other (16.8%), Science-related Non-research (15.3%), Industry Research (14.5%), Academia: Postdoc (10.7%) and Non-science-related (4%); we were unable to determine the career path of the remaining 10.9% of alumni. While positions in Academia (Principal Investigator, Postdoc and Other) remained the most common destination for more recent alumni, entry into Science-related Non-research, Industry Research and Non-science-related positions has increased over time, and entry into Academia: Principal Investigator positions has decreased. Our analysis also reveals information on a number of factors – including publication records – that correlate with the career paths followed by researchers.
Introduction
Career paths in the life sciences have changed dramatically in recent decades, partly because the number of early-career researchers seeking permanent research positions has continued to significantly exceed the number of positions available ( Cyranoski et al., 2011 ; Schillebeeckx et al., 2013 ). Other changes have included efforts to improve research culture, growing concerns about mental health ( Evans et al., 2018 ; Levecque et al., 2017 ), increased collaboration ( Vermeulen et al., 2013 ), an increased proportion of project-based funding ( Lepori et al., 2007 ; Jonkers and Zacharewicz, 2016 ) and greater awareness of careers outside academic research ( Hayter and Parker, 2019 ). Nevertheless, many PhD students and postdocs remain keen to pursue careers in research and, if possible, secure a permanent position as a Principal Investigator (PI) at a university or research institute ( Fuhrmann et al., 2011 ; Gibbs et al., 2015 ; Lambert et al., 2020 ; Roach and Sauermann, 2017 ; Sauermann and Roach, 2012 ).
Data on career paths in the life sciences have become increasingly available in recent years ( Blank et al., 2017 ; Council for Doctoral Education, 2020 ), and such data are useful to individuals as they plan their careers, and also to funding agencies and institutions as they plan for the future. In this article we report the results of a time-resolved analysis of the career paths of 2284 researchers who completed a PhD or postdoc at the European Molecular Biology Laboratory ( EMBL ) between 1997 and 2020. This period included major global events, such as financial crisis of 2007 and 2008 ( Izsak et al., 2013 ; Pellens et al., 2018 ), and also major events within the life sciences (such as the budget of the US National Institutes of Health doubling between 1998 and 2003 and then plateauing; Wadman, 2012 ; Zerhouni, 2006 ).
EMBL is an intergovernmental organisation with six sites in Europe, and its missions include scientific training, basic research in the life sciences, and the development and provision of a range of scientific services. The organization currently employs more than 1110 scientists, including over 200 PhD students, 240 postdoctoral fellows, and 80 PIs. EMBL has a long history of training PhD students and postdocs, and the EMBL International PhD Programme – one of the first structured PhD programmes in Europe – has a completion rate of 92%, with students taking an average of 3.95 years to submit their thesis (data for 2015–2019). More recently, EMBL has launched dedicated fellowship programmes with structured training curricula for postdocs.
Data collection for this study was initially carried out in 2017 and updated in 2021. Using manual Google searches, we located publicly available information identifying the current role of 89% (2035/2284) of the sample ( Table 1 ). These alumni were predominantly based in the European Union (60%, 1224/2035), other European countries including UK and Switzerland (20%), and the US (11%). For 71% of alumni (1626/2284), we were able to reconstruct a detailed career path based on online CVs and biographies (see Methods). EMBL alumni also ended up in a range of careers, which were classified as follows: Academia: Principal Investigator; Academia: Postdoc; Academia: Other research/teaching/service role; Industry Research; Science-related Non-research; and Non-science-related. We also collected data on different types of jobs within the last three of these career areas ( Table 2 ).
See Table 2 for more information on the different jobs covered by Industry Research, Science-related Non-research, and Non-science-related. This classification is based on Stayart et al., 2020 .
AcPI: includes those leading an academic research team with financial and scientific independence – evidenced by a job title such as group leader, professor, associate professor or tenure-track assistant professor. Where the status was unclear from the job title, we classified an alumnus as a Principal Investigator (PI) if one of the following criteria was fulfilled: (a) they appear to directly supervise students/postdocs (based on hierarchy shown on website); (b) they have published a last author publication from their current position; (c) their group website or CV indicates that they have a grant (not just a personal merit fellowship) as a principal investigator. AcOt: differs from Stayart et al., 2020 in that it includes academic research, scientific services or teaching staff (e.g., research staff, teaching faculty and staff, technical directors, research infrastructure engineers).
On average, the alumni in our sample published an average of 4.5 research articles about their work at EMBL, and were the first author on an average of 1.6 of those articles (Table S1 in Supplementary file 1 ). Overall, 90% of the sample (2047/2284) authored at least one article about their EMBL work, and 73% (1666/2284) were the first author on at least one article. The average time between being awarded a PhD and taking up a first role in a specific career area ranged from 4.2 years for Non-science-related positions to 6.8 years for a Principal Investigator (PI) position.
Most alumni remain in science
The majority of alumni (1263/2284=55.3%) were found to be working in an academic position in 2021, including 636 who were PIs, 244 who were in Academia: Postdoc positions, and 383 who were working in Academia: Other positions, which included teaching, research and working for a core facility/technology platform ( Figure 1A ). Just under one-sixth (332/2284=14.5%) were employed in Industry Research positons, and a similar proportion (349/2284=15.3%) were employed in Science-related Non-research positions, such as technology transfer, science administration and education, and corporate roles at life sciences companies. Around 4% were employed in professions not related to science, and the current careers of around 11% of alumni were unknown.

( A ) Charts showing the percentage of PhD alumni (n=969) and postdoc alumni (n=1315) from EMBL in different careers in 2021 (see Table 1 ). ( B ) Charts showing percentage of PhD (left, n=800) and postdoc (right, n=1053) alumni in different careers five years after finishing their PhD or postdoc, for three different cohorts. Chart excludes 169 PhD students and 262 postdocs who have not yet reached the five-year time point. ( C ) Charts showing the percentage of PhD alumni from EMBL (blue column) in PI positions with the percentage of PhD alumni from Stanford University (grey column) in research-focused faculty positions ( Stanford Biosciences, 2021 ). Detailed information about the comparison group can be found in Table S3 in Supplementary file 1 .
Figure 1—source data 1.
Figure 1—figure supplement 1..

( A ) Sankey diagram showing that of the 539 alumni who have held an Academia: PI (AcPI) position at some time, 75.3% moved into their first AcPI position from an Academia: Postdoc (AcPD) position, and 20.6% moved from an Academia: Other (AcOt) positon; 93.1% of these alumni are still in an AcPI position. Abbreviations and percentages for a position are only shown for values of 10% or higher. ( B ) Similar Sankey diagram for the 477 alumni who have held an AcOt position at some time. ( C ) Similar Sankey diagram for the 415 alumni who have held an Industry Research (IndR) position at some time. ( D ) Similar Sankey diagram for the 364 alumni who have held a Science-related Non-research (NonRes) position at some time. ( E ) Similar Sankey diagram for the 131 alumni who have held a Non-science-related (NonSci) position at some time. Data are shown only for alumni for whom a detailed career path is available (n=1626). A preceding AcPD position includes entry direct from an EMBL postdoc and entry via a postdoc position held after leaving EMBL. If an EMBL PhD student became a bridging postdoc in the same lab, this is included in the Academia: PhD category. Diagrams were created in R and scaled manually so that the height is proportional to the number of alumni in the role.
Figure 1—figure supplement 1—source data 1.
Figure 1—figure supplement 2..

Charts showing the percentage of PhD and postdoc alumni in different roles at five different time points for three different cohorts. See Table 3 for cohort sizes; alumni who have not yet reached a given time point are not included. ( A ) PhD alumni 1 year after EMBL (n=969). ( B ) Postdoc alumni 1 year after EMBL (n=1315). ( C ) PhD alumni 5 years after EMBL (n=800). ( D ) Postdoc alumni 5 years after EMBL (n=1053). ( E ) PhD alumni 9 years after EMBL (n=597). ( F ) Postdoc alumni 9 years after EMBL (n=791). ( G ) PhD alumni 13 years after EMBL (n=419). ( H ) Postdoc alumni 13 years after EMBL (n=578). ( I ) PhD alumni 17 years after EMBL (n=256). ( J ) Postdoc alumni 17 years after EMBL (n=369).
Figure 1—figure supplement 2—source data 1.
Figure 1—figure supplement 3..

Charts comparing percentage of PhD alumni in PI or PI-like positions for EMBL and Stanford University, the University of California San Francisco (UCSF), the University of Chicago (Bioscience Division), the University of Michigan, and the University of Toronto (Life Sciences Division) for different cohorts at various time points. ( A ) Chart comparing PhD alumni from EMBL in PI positions (blue) and Stanford in tenure-track faculty positions (grey) for two cohorts; the Stanford data were collated in 2013 ( Stanford IT&DS, 2020 ). ( B ) Chart comparing PhD alumni from EMBL in PI positions (blue) and PhD alumni from UCSF in tenure-track faculty positions (grey) a 2002–2006 cohort after 10 years and 5 years, and a 2007–2011 cohort after 5 years ( UCSF Graduate Division, 2021 ) ( C ) Chart comparing PhD alumni from EMBL in PI positions (blue) and PhD alumni from Chicago in tenure-track faculty positions (grey) for two cohorts ( University of Chicago, 2021 ). ( D ) Chart comparing PhD alumni from EMBL in PI positions (blue) and PhD alumni from Michigan in tenure-track faculty positions (grey) for two cohorts. ( University of Michigan, 2018 ) ( E ) Chart comparing PhD alumni from EMBL in PI positions (blue) and PhD alumni from Toronto in tenure-track faculty positions (grey) for three cohorts; the Torotono data were collated in 2016 (University of Toronto, no date). Detailed information about the comparison groups, including cohort sizes, can be found in Table S3 in Supplementary file 1 ; percentages are the number alumni known to be in a PI or PI-like position at the relevant time point as a percentage of all PhD students in that cohort (including students whose position was unknown at that time point).
Of those who became PIs, 75.3% moved from a postdoc to their first PI position, with 20.6% moving from an Academia: Other position ( Figure 1—figure supplement 1A ). On average, PhD alumni became PIs 6.1 calendar years after their PhD defence, and postdoc alumni became PIs 2.5 years after completing their EMBL postdoc. Almost half of the postdoc alumni who became PIs did so directly after completing their EMBL postdoc (168 of 343). Other postdoc alumni made the transition later, most frequently after one additional postdoc (71 alumni) or a single Academia: Other position (56 alumni). 40 alumni held multiple academic positions between their EMBL postdoc and their first PI position, and eight had one or more non-academic positions during this period.
The career paths of those in other positions were more varied ( Figure 1—figure supplement 1B–E ). For example, for alumni who moved into Industry Research, 20.2% entered their first industry role directly from their PhD, 56.4% from a postdoc position, and 13.3% from Academia: Other positions. Moreover, 71.6% remained in this type of role long-term.
The wide variation in job titles used outside academia makes it difficult to assess career progression, but almost 60% (453/766) of alumni working outside academia had a current job title that included a term indicative of a management-level role (such as manager, leader, senior, head, principal, director, president or chief). For leavers from the last five years (2016–2020), this number was 45% (78/174), suggesting that a large proportion of the alumni who leave academia enter – or are quickly promoted to – managerial positions.
For further analysis, EMBL alumni were split into three 8 year cohorts. More recent cohorts were larger, reflecting the growth of the organization between 1997 and 2020, and also contained a higher percentage of female researchers ( Table 3 ). When comparing cohorts, we observed some differences in the specific jobs being done by alumni outside academia 2021 (Table S2 in Supplementary file 1 ). For example, the percentage of alumni involved in ‘data science, analytics, software engineering’ roles increased from 2% (11/625) for the 1997–2004 cohort to 4% (37/896) for the 2013–2020 cohort. However, the absolute number of alumni for most jobs outside academia was small, so our time-resolved analysis therefore focussed on the broader career areas described above.
Percentage of EMBL alumni who become PIs is similar to that for other institutions
For all timepoints, the percentages of alumni from the 2005–2012 and 2013–2020 cohorts working in PI positions in 2021 were lower than the percentage for the 1997–2004 cohort ( Figure 1—figure supplement 2 ). To assess whether this pattern was specific to EMBL, we compared our data with data from other institutions, noting that different institutions can use different methods to collect data and classify career outcomes. We also note that career outcomes are influenced by the broader scientific ecosystem and by the subject focus of institutions and departments, which may attract early-career researchers with dissimilar career motivations. Nevertheless, comparing long-term outcomes with other institutions allows us to interrogate whether the changes we observe for the most frequent, well-defined and linear career path – the PhD–>Postdoc–>PI career path – reflect a general trend.
A number of institutions have released data on career outcomes for PhD students. Stanford University, for example, has published data on the careers of researchers who received a PhD between 2000 and 2019 ( Stanford Biosciences, 2021 ): Stanford has reported that 34% (145/426) of its 2000–2005 PhD alumni were in research-focussed faculty roles in 2018, and that 13% (63/503) of its 2011–2015 PhD alumni were in PI roles; these numbers are comparable to the figures of 37% (78/210) and 11% (25/234) we observe for EMBL alumni for the same time periods ( Figure 1C ). The EMBL data are also comparable to data from the life science division at the University of Toronto ( Reithmeier et al., 2019 ; University Toronto, 2021 ): for example, Toronto has reported that 31% (192/629) of its 2000–2003 graduates and 25% (203/816) of its 2004–2007 graduates were in tenure stream roles in 2016; the corresponding figures for EMBL were 39% (52/132) and 28% (49/172).
We also compared our EMBL data with data from the University of Michigan, the University of California at San Francisco, and the University of Chicago, and found similar proportions of alumni entering PI positions for comparable cohorts ( Figure 1—figure supplement 3 ). This is consistent with our hypothesis that the differences between cohorts are not EMBL-specific, and reflect a wide-spread change in the number of PhDs and postdocs relative to available PI positions.
We did not analyse the data for other career outcomes, as the smaller numbers of individuals in these careers made it difficult to identify real trends. Moreover, only a small number of institutions have released detailed data on the career destinations of recent postdoc alumni, and we are not aware of any long-term cohort-based data.
The proportion of EMBL alumni who become PIs has decreased with time
To estimate the probability of alumni from different cohorts entering a specific career each year after completing a PhD or postdoc at EMBL, we fitted the data to a Cox proportional hazards model. This is a statistical regression method that is commonly used to model time-to-event distributions from observational data with censoring (i.e., when not all study subjects are monitored until the event occurs, or the event never occurs for some of the subjects). In brief, we fitted the data to a univariate Cox proportional hazards model to calculate hazard ratios, which represent the relative chance of the event considered (here: entering a specific career) occurring in each cohort with respect to the oldest cohort. We also calculated Kaplan–Meier estimators, which estimate the probability of the event (entering a specific career) at different timepoints.
For both PhD and postdoc alumni entering PI positions, we observe hazard ratios of less than one in the Cox models when comparing the newer cohorts with the oldest cohort (Table S4 in Supplementary file 1 ), which indicates that the chances of becoming a PI have become lower for the newer cohorts. The Kaplan–Meier plots illustrate lower percentages of PIs among alumni from the most recent cohorts compared to the oldest cohort at equivalent timepoints ( Figure 2A ). Nevertheless, becoming a PI remained the most common career path for alumni from the 2005–2012 cohort (90/341=26.4% for PhD alumni) and (123/422=29.1% for postdoc alumni), and the most recent cohort of alumni appear to be on a similar trajectory.

( A ) Kaplan–Meier plots showing the estimated probability of an individual being in a PI position (y-axis) as a function of time after EMBL (x-axis) for three cohorts of PhD alumni (left) and three cohorts of postdoc alumni (right). Time after EMBL refers to the number of calendar years between PhD defence or leaving the EMBL postdoc programme and first PI position. ( B–E ) Similar Kaplan–Meier plots for Academia: Other positions ( B ), Industry Research positions ( C ), Science-related Non-research positions ( D ), and Non-science-related professions ( E ). Hazard ratios calculated by a Cox regression model can be found in Table S4 in Supplementary file 1 .
Figure 2—figure supplement 1.

( A ) Box plot with overlaid dot plot showing the distribution of the length of time between PhD and first PI role for two cohorts of alumni who defended their PhD between 1997 and 2012 and became a PI within nine calendar years (and for whom we have a detailed career path; n=157). The mean value is indicated as a red cross and the p-values were calculated using Welch’s t-test. The difference between the mean for the two cohorts (5.2 years and 6.1 years) was statistically significant ( P =0.01496). ( B ) Plots for length of time between completion of an EMBL postdoc and first PI role for two cohorts of alumni who completed their postdoc between 1997 and 2012 (n=218). The difference between the two cohorts was not statistically significant ( P =0.192). ( C ) Plots for length of time between PhD and first PI role for the two cohorts of alumni who completed their EMBL postdoc between 1997 and 2012 (and for whom we know year of PhD and have a detailed career path; n=146; note these alumni completed their PhD somewhere else before starting a postdoc at EMBL). The difference between the mean for the two cohorts (5.3 years and 6.0 years) was statistically significant ( P =0.0325).
Figure 2—figure supplement 1—source data 1.
Kaplan–Meier plots show increased proportions of the 2005–2012 and 2013–2020 cohorts entering Science-related Non-research and Non-science-related positions, compared to the 1997–2004 cohort for both PhD and postdoc alumni ( Figure 2D, E ). For the most recent (2013–2020) cohort, there was also an increased rate of entry into Industry: Research positions compared to alumni from PhD and postdoc cohorts from 1997 to 2004 and 2005–2012 ( Figure 2C , Table S4 in Supplementary file 1 ). For Academia: other positions, the rate of entry was similar for all three PhD cohorts, though some differences between cohorts were observed for postdoc alumni ( Figure 2B ).
A small increase in time between year of PhD and first PI position
We decided to explore to what extent increasing postdoc length may contribute to the decreased proportion of alumni who are found as PIs in the years after leaving EMBL. In order to fairly compare alumni from different cohorts, we included only alumni for whom we had a detailed career path, who had defended their PhD at least nine years ago, and who had become a PI within nine years of defending their PhD. We chose a nine-year cut-off because this was the time interval between the last PhDs in the 2005–2012 cohort and the execution of this study; moreover, for PhD alumni from the oldest cohort (1997–2004), most of those who became PIs had done so within nine years (89/97=92%).
157 of the PhD alumni in our sample met these criteria, taking an average of 5.6 calendar years to become a PI (see Methods). There was a statistically significant difference in the average time from PhD to first PI position between the 1997–2004 cohort (5.2 years) and the 2005–2012 cohort (6.1 years; Figure 2—figure supplement 1A ). 218 of the postdoc alumni in our sample met these criteria, taking an average of 2.5 calendar years to become a PI after leaving EMBL (see Methods). There was no statistically significant difference in time between EMBL and first PI role for the 1997–2004 and 2005–2012 postdoc cohorts ( Figure 2—figure supplement 1B ). However, the time between receiving their PhD and becoming a PI increased by from 5.3 calendar years for the 1997–2004 postdoc cohort to 6.0 calendar years for the 2005–2012 postdoc cohort ( Figure 2—figure supplement 1C ).
Gender differences in career outcomes
Many studies have reported that female early-career researchers are less likely to remain in academia ( Alper, 1993 ; Martinez et al., 2007 ). Consistent with these studies, male alumni from EMBL were more likely than female alumni to end up in a PI position ( Figure 3A and B ; Table S5 in Supplementary file 1 ). However, for alumni from 1997 to 2012, there was no statistically significant difference in the length of time taken by male and female alumni to become PIs ( Figure 3—figure supplement 1 ). Female alumni were more likely to end up in a Science-related Non-research position, and male alumni were more likely to end up in an Industry Research or Non-science-related position ( Figure 3 ; Figure 3—figure supplement 2 ). However, female alumni were also more likely to be classified as unknown, and since it is more difficult to follow careers outside the academic world, it is possible that the number of women who established careers outside academia (in positions such as Industry Research, Science-related Non-research, and Non-science-related) is higher than our results suggest.

( A ) Charts showing the percentage of female (n=415) and male (n=554) PhD alumni, and female (n=492) and male (n=823) postdoc alumni, in different careers in 2021. ( B ) Kaplan–Meier plots showing the estimated probability of an individual being in a PI position (y-axis) as a function of time after EMBL (x-axis), stratified by gender for PhD alumni (left) and postdoc alumni (right). ( C ) Kaplan–Meier plots showing the estimated probability of an individual being in a science-related non-research position as a function of time after EMBL, stratified by gender for PhD alumni (left) and postdoc alumni (right). Kaplan–Meier plots for other career outcomes are shown in Figure 3—figure supplement 2 . Hazard ratios calculated by a Cox regression model can be found in Table S5 in Supplementary file 1 .
Figure 3—source data 1.
Figure 3—figure supplement 1..

( A ) Box plot with overlaid dot plot showing the distribution of the length of time between PhD and first PI role for female alumni (left) and male alumni (right) who defended their PhD between 1997 and 2012 and became a PI within nine calendar years (and for whom we have a detailed career path; n=157). The mean value is indicated as a red cross and the p-values calculated using Welch’s t-test. The difference between the mean values for female and male alumni (6.1 years and 5.4 years) was not statistically significant ( P =0.0719). ( B ) Plots for length of time between completion of an EMBL postdoc and first PI role for female alumni (left) and male alumni (right) who completed their postdoc between 1997 and 2012 (n=218). The difference between the mean values for female and male alumni (2.3 years and 3.1 years) was not statistically significant ( P =0.0596). ( C ) Plots for length of time between PhD and first PI role for female alumni (left) and male alumni (right) who completed their EMBL postdoc between 1997 and 2012 and for whom we know year of PhD (and for whom we have a detailed career path; n=146). The difference between the mean values for female and male alumni (5.6 years and 5.7 years) was not statistically significant ( P =0.778).
Figure 3—figure supplement 1—source data 1.
Figure 3—figure supplement 2..

( A ) Kaplan–Meier plots showing the estimated probability of an individual working in Academia: Other (y-axis) as a function of time after EMBL, stratified by gender for PhD alumni (left) and postdoc alumni (right). ( B, C ) Similar Kaplan–Meier plots for alumni working in Industry Research ( B ) and in Non-science-related careers ( C ). Hazard ratios calculated by a Cox regression model can be found in Table S5 in Supplementary file 1 .
Future PIs, on average, published more papers while at EMBL
Publication metrics have been linked to the likelihood of obtaining ( van Dijk et al., 2014 ; Tregellas et al., 2018 ) and succeeding ( von Bartheld et al., 2015 ) in a faculty position. In this study, alumni who became PIs had more favourable publication metrics from their EMBL work – for example, they published more articles, and their papers had higher CNCI values. (CNCI is short for Category Normalized Citation Impact, and a CNCI value of one means that the number of citations received was the same as the average for other articles in that field published in the same year; Figure 4A and B ; Table S6 in Supplementary file 1 ). Using univariate Cox models for time to PI as a function of number of first-author research articles from EMBL work, we estimated that a postdoc with one first-author publication was 3.2 times more likely to be found in a PI position than a postdoc without a first-author publication (95% confidence interval [2.2, 4.7]), and a post-doc with two or more first-author publications was 6.6 times more likely (95% confidence interval [4.7, 9.3]; Figure 4C ).

( A ) Histograms showing the number of alumni who have 0, 1, 2, 3,... first-author articles from their time at EMBL and became PIs (bottom; n=662, excluding 23 outliers), and did not become PIs (top; n=1594, excluding 5 outliers). For clearer visualization, and to protect the identity of alumni with outlying numbers of publications, the x-axis is truncated at the 97.5 th percentile. The mean for each group (including outliers) is shown as a red dashed line; alumni who became PIs have an average of 2.4 first-author articles from their time at EMBL, whereas other alumni have an average of 1.2 articles; this difference is significant ( P <2.2 × 10 –16 ; Welch’s t-test). ( B ) 1656 alumni had one or more first-author articles from their time at EMBL that had a CNCI value in the InCites database. For each of these alumni, the natural logarithm of the highest CNCI value was calculated, and these histograms show the number of alumni for which this natural logarithm is between –4.5 and –3.5, between –3.5 and –2.5, and so on; the bottom histogram is for alumni who became PIs, and the top histogram is for other alumni. A CNCI value of 1 (plotted here at ln(1)=0; vertical black line) means that the number of citations received by the article is the same as the average for other articles in that field published in the same year. The mean for each group is shown as a red dashed line; alumni who became PIs have an average CNCI of 5.7, whereas other alumni have an average CNCI of 3.1; this difference is significant ( P <2.829 × 10 –6 ; Welch’s t-test). ( C ) Kaplan–Meier plots showing the estimated probability of an individual becoming a PI (y-axis) as a function of time after EMBL (x-axis), stratified by number of first-author publications from research completed at EMBL, for PhD alumni (left) and postdoc alumni (right). Hazard ratios calculated by a Cox regression model can be found in Table S7 in Supplementary file 1 . ( D ) Harrell’s C-Index for various Cox models for predicting entry into PI positions. The first seven bars show the C-index for univariate and multivariate models for a subset of covariates (which subset is shown below the x-axis), and the eighth bar is for a multivariate model that includes the covariates from all subsets. The subsets are time & cohort (multivariate, including the variables: cohort, PhD year (if known), start and end year at EMBL), predoc (ie PhD student)/postdoc (univariate), group leader seniority (univariate), nationality (univariate), gender (univariate), publications (multivariate: containing variables related to the alumni’s publications from their EMBL work; these are variables with a name beginning with “pubs” in Table S1 in Supplementary file 1 ) and group publications (multivariate: containing variables related to the aggregated publication statistics for all PhD students and postdocs who were trained in the same group; these are variables with a name beginning with “group_pubs” in Table S1 in Supplementary file 1 ). A value of above 0.5 indicates that a model has predictive power, with a value of 1.0 indicating complete concordance between predicted and observed order to outcome (e.g. entry into a PI position). Bars denote the mean, and the error bars show the 95% confidence intervals. A value of above 0.5 indicates that a model has predictive power, with a value of 1.0 indicating complete concordance between predicted and observed order to outcome (e.g. entry into a PI position). Bars denote the mean, and the error bars show the 95% confidence intervals.
Figure 4—source data 1.
Figure 4—figure supplement 1..

Harrell’s C-Index for various Cox models for predicting entry into Academia: Other ( A ), Industry Research ( B ), Science-related Non-research ( C ), and Non-science-related careers ( D ). As in Figure 4D , the first seven bars show the C-index for univariate and multivariate models containing subsets of variables, the eighth bar is for a multivariate model containing all variables, and a value of above 0.5 indicates that a model has predictive power.
Figure 4—figure supplement 1—source data 1.
Figure 4—figure supplement 2..

Kaplan–Meier plots showing the estimated probability of an individual being in various careers (y-axis) as a function of time after EMBL (x-axis), stratified by number of first-author publications from research completed at EMBL for PhD alumni (left) and postdoc alumni (right). Hazard ratios calculated by a Cox regression model can be found in Table S7 in Supplementary file 1 .
Publication factors are highly predictive of entry into a PI position
To understand the potential contribution of publication record in the context of other factors – including cohort, gender, nationality, publications, and seniority of the supervising PI – we fitted multivariate Cox models. To quantify publication record, we considered a range of metrics including journal impact factor, which has been shown to statistically correlate with becoming a PI in some studies ( van Dijk et al., 2014 ) and has been used by some institutions in research evaluation ( McKiernan et al., 2019 ). It should be stressed, however, that EMBL does not use journal impact factors in hiring or evaluation decisions, and is a signatory of the San Francisco Declaration on Research Assessment (DORA) and a member of the Coalition for Advancing Research Assessment (CoARA).
To evaluate the predictive power of each Cox model, we used the cross-validated Harrell’s C-index, which measures predictive power as the average agreement across all pairs of individuals between observed and predicted temporal order of the outcome (in our case, entering a specific type of position; see Methods). A C-index of 1 indicates complete concordance between observed and predicted order. For example, for a model of entry into PI roles, a C-index of 1 would mean that the model correctly predicts, for all pairs of individuals, which individual becomes a PI first based on the factors included in the model. A C-index 0.5 is the baseline that corresponds to random guessing. Prediction is clearly limited by the fact that we could not explicitly encode some covariates that are certain to play an important role in career outcomes, such as career preferences and relevant skills. Nevertheless, the C-index for models containing all data were between 0.61 (entry to Industry Research, Figure 4—figure supplement 1B ) and 0.70 (entry into PI positions, Figure 4D ), suggesting that the factors have some predictive power.
To investigate which factors were most predictive for entry into different careers, we compared models containing different sets of factors. Consistent with previous studies, we found that statistics related to publications were highly predictive for entry into a PI position: a multivariate model containing only the publication statistics performs almost as well as the complete multivariate model, reaching a C-index of 0.69 ( Figure 4D ). The publications of the research group the alumnus was trained in (judged by the aggregated publication statistics for all PhD students and postdocs who were trained in the same group) was also predictive, with a C-index of 0.61.
Cohort/year, gender, and status at EMBL (PhD or postdoc) were also predictors of entry into a PI position in our Cox models, with C-indexes of 0.59, 0.57 and 0.55, respectively ( Figure 4D ). This is consistent with our observation that alumni from earlier cohorts/years ( Figure 1B ), male alumni ( Figure 3A ) and postdoc alumni ( Figure 1A ) were more frequently found in PI positions. Models containing only nationality or group leader seniority were not predictive.
For Academia: Other positions, the factors that were most predictive were those related to publications of the research group the alumnus was trained in ( Figure 4—figure supplement 1A ). It is unclear why this might be, but we speculate that this could reflect publication characteristics specific to certain fields that have a high number of staff positions, or other factors such as the scientific reputation, breadth or collaborative nature of the research group and its supervisor. The group’s publications were also predictive for Industry Research and Science-related Non-research positions.
Time-related factors (i.e., cohort, PhD award year and EMBL contract start/end years) were the strongest prediction factors for Industry Research, Science-related Non-research, and Non-science-related positions ( Figure 4—figure supplement 1B–D ), and more recent alumni were more frequently found in these careers ( Figure 2C–E ).
Overall, statistics related to an individual’s own publications were a weak predictor for entry into positions other than being a PI ( Figure 4—figure supplement 1 ; Figure 4—figure supplement 2 ; Table S7–S11 in Supplementary file 1 ). For example, for Industry Research, a model containing statistics for an individual’s publications had a C-index of only 0.53, compared to 0.61 for the complete model, and there were no differences in likelihood of a PhD alumnus with 0, 1 or 2+publications entering an Industry Research position.
Changes in the publications landscape
Reports suggest that the number of authors on a typical research article in biology has increased over time, as has the amount of data in a typical article ( Vale, 2015 ; Fanelli and Larivière, 2016 ); a corresponding decrease in the number of first-author research articles per early-career researcher has also been reported ( Kendal et al., 2022 ). For articles linked to the PhD students and postdocs in this study, the mean number of authors per article has more than doubled between 1995 and 2020 ( Figure 5A ). The mean number of articles per researcher did not change between the three cohorts studied ( Figure 5B ; the mean was 3.6 articles per researcher), but researchers from the second and third cohorts published fewer first-author articles than those from the first cohorts ( Figure 5C ). However, more recent articles had higher CNCI values ( Figure 5D ). The proportion of EMBL articles that included international collaborators also increased from 47% in 1995 to 79% in 2020.

( A ) Mean number of authors (y-axis) as a function of year (x-axis) for research articles that were published between 1995 and 2020, and have at least one of the alumni included in this study as an author (n=5413); the winsorized mean has been used to limit the effect of outliers. The mean number of authors has increased by a factor of more than two between 1995 and 2000. ( B ) Boxplot showing the distribution of the number of articles published per researcher for three cohorts. The mean is indicated as a red cross; the circles are outliers. No statistically significant difference was found between the cohorts; the p-value of 0.1156 was generated using a one-way analysis of variance (ANOVA) test of the full dataset (including outliers); the p-value excluding outliers is 0.26. ( C ) Boxplot showing the distribution of the number of first-author articles published per researcher for three cohorts; the two most recent cohorts published fewer first-author articles than the 1997–2004 cohort; the p-value (excluding outliers) was 6.5x10 –7 ; see Table S12 in Supplementary file 1 . ( D ) Mean CNCI (y-axis) as a function of time (x-axis) for research articles that were published between 1995 and 2020, and have at least one of the alumni included in this study as an author (n=5413). Recent articles have higher CNCI values. For clearer visualization, and to protect the identity of alumni with outlying numbers of publications, the y-axis in ( B ) and ( C ) is truncated at the 97.5 percentile.
Figure 5—source data 1.
Many early-career researchers are employed on fixed-term contracts funded by project-based grants, sometimes for a decade or more ( OECD, 2021 ; Acton et al., 2019 ), and surveys suggest that early-career researchers are concerned about career progression ( Woolston, 2020 ; Woolston, 2019 ). We hope PhD students and postdocs will be reassured to learn that the skills and knowledge they acquire during their training are useful in a range of careers both inside and outside acaemica.
Further changes to the career landscape in the life sciences are likely in future, not least as a result of the long-term impacts of the COVID-19 pandemic ( Bodin, 2020 ). It is essential, therefore, that early-career researchers are provided with opportunities to reflect on their strengths, to understand the wide range of career options available to them, and to develop new skills.
The provision of effective support for PhD students and postdocs will require input from different stakeholders – including funders, employers, supervisors and policy makers – and the engagement of the early-career researchers themselves. At EMBL, a career service was launched in 2019 for all PhD students and postdocs, building on a successful EC-funded pilot project that offered career support to 76 postdocs in the EMBL Interdisciplinary Postdoc Programme. The EMBL Fellows’ Career Service now offers career webinars and a blog to the whole scientific community as well as additional tailored support for EMBL PhDs and postdocs including individual career guidance, workshops, resources and events. Funders and policymakers may also need to reassess the sustainably of academic career paths, and to review how funding is allocated between project-based grants and mechanisms that can support PI and non-PI positions with longer-term stability. These measures will will also support equality, diversity and inclusion in science, particularly if paired with research assessment practices that consider factors that can affect apparent research productivity such as career breaks, teaching and service activities.
Factors related to publication are highly predictive of entry into PI careers, and one challenge for an early-career researcher hoping to pursue such a career is to balance the number of articles they publish with the subjective quality of these articles. The trend towards fewer first-author articles per researcher likely reflects a global trend towards articles with more authors and a greater focus on collaborative and/or interdisciplinary approaches to research. Working on a project that involves multiple partners provides an early-career researcher with the opportunity to develop a range of skills, including teamwork, leadership and creativity. Such projects also allow researchers to tackle challenging biological questions from new angles to advance in their field of research, something viewed very positively by academic hiring committees ( Hsu et al., 2021 ; Clement et al., 2020 ; Fernandes et al., 2020 ); however, multi-partner interdisciplinary projects can also take longer to complete. It is therefore important that early-career researchers and their supervisors discuss the potential impact and challenges of (prospective) projects, and what can be done to reduce any risks. For example, open science practices – including author credit statements, FAIR data, and pre-printing – can make project contributions more transparent and available faster ( Kaiser, 2017 ; McNutt et al., 2018 ; Wilkinson et al., 2016 ; Wolf et al., 2021 ).
Limitations
The limitations of our study include that its retrospective, observational design limits our ability to disentangle causation from correlation. The changes in career outcomes may be driven primarily by increased competition for PI roles, but they could also be influenced by a greater availability or awareness of other career options. EMBL has held an annual career day highlighting career options outside academia since 2006, and many of our alumni decide to pursue a career in the private sector, attracted by perceptions of higher pay, more stable contracts, and/or better work-life balance. Likewise, early-career researchers with an interest in a specific technology might, for example, prefer to work at a core facility.
Additionally, we cannot exclude the possibility that other factors may also affect the differences we see between cohorts (such as variations in the number of alumni taking up academic positions in countries that offer later scientific independence). Finally, although comparisons with data from the US and Canada suggest that the trend towards fewer alumni becoming PIs is a global phenomenon, it is possible that some of the trends we observe are specific to EMBL.
We plan to update our observational data every four years, and to maintain data on the career paths of alumni for 24 years after they leave EMBL. This will help us to identify any further changes in the career landscape and to better understand long-term career outcomes in the life sciences. Silva et al., 2019 have also described a method for tracking career outcomes on a yearly basis with estimations of the time and other resources required. We encourage institutions to consider whether they can adapt our methods, or Silva’s method, to the administrative processes and data-privacy regulations applicable at their institutions.
Future studies should also ideally include mixed-method longitudinal studies, which would allow information on career motivations, skills development, research environment, job application activity and other factors to be recorded. Combining the results of such studies with data on career outcomes would allow multifactorial and complex issues, such as gender differences in career outcomes, to be investigated, and would also provide policymakers with a fuller picture of workforce trends. Such studies would, however, require multiple institutions to commit to supplying large amounts of data every year, and coordinating the collection and analysis of such data year-on-year would be a major undertaking that would require the support of funders and institutions.
Data collection and analysis
The study includes individuals who graduated from the EMBL International PhD Programme between 1997 and 2020 (n=969), or who left the EMBL postdoc programme between 1997 and 2020 after spending at least one year as an EMBL postdoctoral fellow (n=1315). Each person is included only once in the study: where a PhD student remained at EMBL for a bridging or longer postdoc, they were included as PhD alumni only, with the postdoc position listed as a career outcome.
For each alumnus or alumna, we retrieved demographic information from our internal records and identified publicly available information about each person’s career path (see Supplementary file 2 ). Where possible, this information was used to reconstruct a detailed career path. An individual was classified as having a "detailed career path" if an online CV or biosketch was found that accounted for their time since EMBL excluding a maximum of two one-calendar-year career breaks (which may, for example, reflect undisclosed sabbaticals or parental leave). Each position was classified using a detailed taxonomy, based on a published schema ( Stayart et al., 2020 ), and given a broad overall classification (see Supplementary file 2 ). The country of the position was also recorded. For the most recent position, we noted whether the job title was indicative of a senior or management level role (i.e., if it included "VP"; "chief"; "cso";"cto"; "ceo"; "head"; "principal”; "president"; "manager"; "leader"; "senior"), or if they appeared to be running a scientific service or core facility in academia.
We use calendar years for all outcome data – for example, for an individual who left EMBL in 2012, the position one calendar year after EMBL would be the position held in 2013. If multiple positions were held in that year, we take the most recent position. We use calendar years, as the available online information often only provides the start and end year of a position (rather than exact date).
An EMBL publication record was also reconstituted for each person in the study. Each of their publications linked to EMBL in the Web of Science and InCites databases in June 2021 were recorded. The data included publication year and – for those indexed in InCites – crude metrics, such as CNCI, percentile in subject area, and journal impact factor. EMBL publications were assigned to individuals in the study based on matching name and publication year (see Supplementary file 2 for full description). When an individual was the second author on a publication, we manually checked for declarations of co-first authorship. Aggregate publication statistics for individuals with the same primary supervisor were also calculated.
The names and other demographic information that would allow easy identification of individuals in the case of a data breach were pseudonymised. A file with key data for analysis and visualisation in R was then generated. A description of this data table can be found in Table S1 in Supplementary file 1 , along with summary statistics.
Statistical model
A Cox proportional hazards regression model was fitted to the data in order to predict time-to-event probabilities for each type of career outcome based on different covariates including cohort, publication variables and gender. Multivariate Cox models were fitted using a ridge penalty with penalty parameter chosen by 10-fold cross-validation. Harrell’s C-index was calculated for each fit in an outer cross-validation scheme for validation and analysis of different models, with 10-fold cross-validation.
Acknowledgements
We thank Monika Lachner and Anne Ephrussi for their critical reading of the manuscript and strong support of this project. We also acknowledge the instrumental support of the Alumni Relations, DPO, HR, SAP, Library, International PhD Programme and Postdoc Programme teams at EMBL. We also thank Edith Heard, Brenda Stride, Jana Watson-Kapps (FMI), and the Directorate, SAC, SSMAC and Council of the EMBL for discussion. The work was supported by: EMBL (JL, BK, MR, WH, RCG) and the EMBL International PhD Programme (BV). RCG is employed by EMBL’s Interdisciplinary Postdoc Programme, which has received funding from the European Union’s Horizon 2020 programme (Marie Skłodowska-Curie Actions).
Biographies
Junyan Lu , Genome Biology Unit, European Molecular Biology Laboratory, Heidelberg, Germany
Britta Velten , Genome Biology Unit, European Molecular Biology Laboratory, Heidelberg, Germany
Bernd Klaus , Genome Biology Unit, European Molecular Biology Laboratory, Heidelberg, Germany
Mauricio Ramm , EMBL International Centre for Advanced Training, European Molecular Biology Laboratory, Heidelberg, Germany
Wolfgang Huber , Genome Biology Unit, European Molecular Biology Laboratory, Heidelberg, Germany
Rachel Coulthard-Graf , EMBL International Centre for Advanced Training, European Molecular Biology Laboratory, Heidelberg, Germany
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Peter Rodgers, eLife United Kingdom .
Funding Information
This paper was supported by the following grants:
- Horizon 2020 Framework Programme 664726 to Rachel Coulthard-Graf.
- Horizon 2020 Framework Programme 847543 to Rachel Coulthard-Graf.
- European Molecular Biology Laboratory to Britta Velten, Bernd Klaus, Mauricio Ramm, Wolfgang Huber, Rachel Coulthard-Graf, Junyan Lu.
Additional information
No competing interests declared.
Data curation, Formal analysis, Visualization, Methodology, Writing – review and editing.
Data curation, Formal analysis, Methodology, Visualization, Writing – review and editing.
Investigation, Methodology.
Supervision, Methodology, Writing – review and editing.
Conceptualization, Data curation, Formal analysis, Investigation, Visualization, Methodology, Writing – original draft, Writing – review and editing.
Additional files
Mdar checklist, supplementary file 1., supplementary file 2., data availability.
- Acton SE, Bell AJ, Toseland CP, Twelvetrees A. A survey of new PIs in the UK. eLife. 2019; 8 :e46827. doi: 10.7554/eLife.46827. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Alper J. The pipeline is leaking women all the way along. Science. 1993; 260 :409–411. doi: 10.1126/science.260.5106.409. [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Blank R, Daniels RJ, Gilliland G, Gutmann A, Hawgood S, Hrabowski FA, Pollack ME, Price V, Reif LR, Schlissel MS. A new data effort to inform career choices in biomedicine. Science. 2017; 358 :1388–1389. doi: 10.1126/science.aar4638. [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Bodin M. University redundancies, furloughs and pay cuts might loom amid the pandemic, survey finds. Nature. 2020 doi: 10.1038/d41586-020-02265-w. [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Clement L, Dorman JB, McGee R. The Academic Career Readiness Assessment: Clarifying hiring and training expectations for future biomedical life sciences faculty. CBE Life Sciences Education. 2020; 19 :ar22. doi: 10.1187/cbe.19-11-0235. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Coulthard R, Lu J. EMBL-Career-Analysis. swh:1:rev:f82a94f925fb44aa59383e0afc167b4f4f0c67bd Software Heritage. 2022 https://archive.softwareheritage.org/swh:1:dir:8e764aae6f23b2d2a6a4ecb3322ada777e604388;origin=https://github.com/Huber-group-EMBL/EMBL-Career-Analysis;visit=swh:1:snp:66c550eefce2ed4462b612ac31ee4d219cc97f6c;anchor=swh:1:rev:f82a94f925fb44aa59383e0afc167b4f4f0c67bd
- Council for Doctoral Education Tracking the Careers of Doctorate Holders. 2020. [September 23, 2023]. https://eua-cde.org/component/attachments/attachments.html?id=2988
- Cyranoski D, Gilbert N, Ledford H, Nayar A, Yahia M. The PhD factory. Nature. 2011; 472 :276–279. doi: 10.1038/472276a. [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Evans TM, Bira L, Gastelum JB, Weiss LT, Vanderford NL. Evidence for a mental health crisis in graduate education. Nature Biotechnology. 2018; 36 :282–284. doi: 10.1038/nbt.4089. [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Fanelli D, Larivière V. Researchers’ individual publication rate has not increased in a century. PLOS ONE. 2016; 11 :e0149504. doi: 10.1371/journal.pone.0149504. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Fernandes JD, Sarabipour S, Smith CT, Niemi NM, Jadavji NM, Kozik AJ, Holehouse AS, Pejaver V, Symmons O, Bisson Filho AW, Haage A. A survey-based analysis of the academic job market. eLife. 2020; 9 :e54097. doi: 10.7554/eLife.54097. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Fuhrmann CN, Halme DG, O’Sullivan PS, Lindstaedt B. Improving graduate education to support a branching career pipeline: Recommendations based on a survey of doctoral students in the basic biomedical sciences. CBE Life Sciences Education. 2011; 10 :239–249. doi: 10.1187/cbe.11-02-0013. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Gibbs KD, McGready J, Griffin K. Career development among American biomedical postdocs. Cell Biology Education. 2015; 14 :ar44. doi: 10.1187/cbe.15-03-0075. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Hayter CS, Parker MA. Factors that influence the transition of university postdocs to non-academic scientific careers: An exploratory study. Research Policy. 2019; 48 :556–570. doi: 10.1016/j.respol.2018.09.009. [ CrossRef ] [ Google Scholar ]
- Hsu NS, Rezai-Zadeh KP, Tennekoon MS, Korn SJ. Myths and facts about getting an academic faculty position in neuroscience. Science Advances. 2021; 7 :eabj2604. doi: 10.1126/sciadv.abj2604. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Izsak K, Markianidou P, Lukach R, Wastyn A. Impact of the Crisis on Research and Innovation Policies. Study for the European Commission DG Research by Technopolis Group Belgium and Idea Consult. 2013. [September 23, 2023]. https://www.technopolis-group.com/wp-content/uploads/2020/02/The-impact-of-the-financial-crisis-on-research-and-innovation-policies-in-EU-Member-States.pdf
- Jonkers K, Zacharewicz T. Research Performance Based Funding Systems: A Comparative Assessment. JRC Publications Repository; 2016. [ CrossRef ] [ Google Scholar ]
- Kaiser J. Are preprints the future of biology? A survival guide for scientists. Science. 2017; 1 :aaq0747. doi: 10.1126/science.aaq0747. [ CrossRef ] [ Google Scholar ]
- Kendal D, Lee KE, Soanes K, Threlfall CG. The great publication race’ vs ‘abandon paper counting’: Benchmarking ECR publication and co-authorship rates over past 50 years to inform research evaluation. F1000Research. 2022; 11 :95. doi: 10.12688/f1000research.75604.1. [ CrossRef ] [ Google Scholar ]
- Lambert WM, Wells MT, Cipriano MF, Sneva JN, Morris JA, Golightly LM. Career choices of underrepresented and female postdocs in the biomedical sciences. eLife. 2020; 9 :e48774. doi: 10.7554/eLife.48774. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Lepori B, van den Besselaar P, Dinges M, Potì B, Reale E, Slipersæter S, Thèves J, van der Meulen B. Comparing the evolution of national research policies: What patterns of change? Science and Public Policy. 2007; 34 :372–388. doi: 10.3152/030234207X234578. [ CrossRef ] [ Google Scholar ]
- Levecque K, Anseel F, De Beuckelaer A, Van der Heyden J, Gisle L. Work organization and mental health problems in PhD students. Research Policy. 2017; 46 :868–879. doi: 10.1016/j.respol.2017.02.008. [ CrossRef ] [ Google Scholar ]
- Martinez ED, Botos J, Dohoney KM, Geiman TM, Kolla SS, Olivera A, Qiu Y, Rayasam GV, Stavreva DA, Cohen-Fix O. Falling off the academic bandwagon. Women are more likely to quit at the postdoc to principal investigator transition. EMBO Reports. 2007; 8 :977–981. doi: 10.1038/sj.embor.7401110. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- McKiernan EC, Schimanski LA, Muñoz Nieves C, Matthias L, Niles MT, Alperin JP. Use of the Journal Impact Factor in academic review, promotion, and tenure evaluations. eLife. 2019; 8 :e47338. doi: 10.7554/eLife.47338. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- McNutt MK, Bradford M, Drazen JM, Hanson B, Howard B, Jamieson KH, Kiermer V, Marcus E, Pope BK, Schekman R, Swaminathan S, Stang PJ, Verma IM. Transparency in authors’ contributions and responsibilities to promote integrity in scientific publication. PNAS. 2018; 115 :2557–2560. doi: 10.1073/pnas.1715374115. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- OECD . Reducing the Precarity of Academic Research Careers. OECD iLibrary; 2021. [ CrossRef ] [ Google Scholar ]
- Pellens M, Peters B, Hud M, Rammer C, Licht G. Public investment in R&D in reaction to economic crises - A longitudinal study for OECD countries. SSRN Electronic Journal. 2018; 1 :3122254. doi: 10.2139/ssrn.3122254. [ CrossRef ] [ Google Scholar ]
- Reithmeier R, O’Leary L, Zhu X, Dales C, Abdulkarim A, Aquil A, Brouillard L, Chang S, Miller S, Shi W, Vu N, Zou C. The 10,000 PhDs project at the University of Toronto: Using employment outcome data to inform graduate education. PLOS ONE. 2019; 14 :e0209898. doi: 10.1371/journal.pone.0209898. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Roach M, Sauermann H. The declining interest in an academic career. PLOS ONE. 2017; 12 :e0184130. doi: 10.1371/journal.pone.0184130. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Sauermann H, Roach M. Science PhD career preferences: Levels, changes, and advisor encouragement. PLOS ONE. 2012; 7 :e36307. doi: 10.1371/journal.pone.0036307. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Schillebeeckx M, Maricque B, Lewis C. The missing piece to changing the university culture. Nature Biotechnology. 2013; 31 :938–941. doi: 10.1038/nbt.2706. [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Silva EA, Mejía AB, Watkins ES. Where do our graduates go? A tool kit for tracking career outcomes of biomedical PhD students and postdoctoral scholars. CBE Life Sciences Education. 2019; 18 :le3. doi: 10.1187/cbe.19-08-0150. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Stanford Biosciences Alumni Career Outcomes by Cohort. 2021. [September 10, 2021]. https://biosciences.stanford.edu/prospective-students/alumni-career-outcomes-dashboard/alumni-career-outcomes-by-cohort/
- Stanford IT&DS The Stanford PhD Alumni Employment Project. 2020. [January 17, 2020]. https://tableau.stanford.edu/t/IRDS/views/StanfordPhDAlumniEmployment/StanfordPhDAlumniEmploymentDashboard?%3Aembed_code_version=3&%3Aembed=y&%3AloadOrderID=0&%3Adisplay_spinner=no&%3Adisplay_count=n&%3AshowVizHome=n&%3Aorigin=viz_share_link
- Stayart CA, Brandt PD, Brown AM, Dahl T, Layton RL, Petrie KA, Flores-Kim EN, Peña CG, Fuhrmann CN, Monsalve GC. Applying inter-rater reliability to improve consistency in classifying PhD career outcomes. F1000Research. 2020; 9 :8. doi: 10.12688/f1000research.21046.2. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Tregellas JR, Smucny J, Rojas DC, Legget KT. Predicting academic career outcomes by predoctoral publication record. PeerJ. 2018; 6 :e5707. doi: 10.7717/peerj.5707. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- UCSF Graduate Division PhD Program Statistics. 2021. [September 9, 2021]. https://graduate.ucsf.edu/program-statistics
- University of Chicago Career Outcomes of PhD Alumni. 2021. [August 20, 2021]. https://biosciences.uchicago.edu/after-uchicago/outcomes
- University of Michigan Rackham Doctoral Program Statistics. 2018. [November 9, 2018]. https://tableau.dsc.umich.edu/#/site/UM-Public/views/RackhamDoctoralProgramStatistics/ProgramStatistics
- University Toronto Employed and Engaged: Career Outcomes of Our PhD Graduates. 2021. [September 10, 2021]. https://www.sgs.utoronto.ca/about/explore-our-data/10000-phds-project/
- Vale RD. Accelerating scientific publication in biology. PNAS. 2015; 112 :13439–13446. doi: 10.1073/pnas.1511912112. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- van Dijk D, Manor O, Carey LB. Publication metrics and success on the academic job market. Current Biology. 2014; 24 :R516–R517. doi: 10.1016/j.cub.2014.04.039. [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Vermeulen N, Parker JN, Penders B. Understanding life together: a brief history of collaboration in biology. Endeavour. 2013; 37 :162–171. doi: 10.1016/j.endeavour.2013.03.001. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- von Bartheld CS, Houmanfar R, Candido A. Prediction of junior faculty success in biomedical research: Comparison of metrics and effects of mentoring programs. PeerJ. 2015; 3 :e1262. doi: 10.7717/peerj.1262. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wadman M. A workforce out of balance. Nature. 2012; 486 :304. doi: 10.1038/486304a. [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wilkinson MD, Dumontier M, Aalbersberg IJJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJG, Groth P, Goble C, Grethe JS, Heringa J, ’t Hoen PAC, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, van Schaik R, Sansone SA, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, van der Lei J, van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, Mons B. The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data. 2016; 3 :160018. doi: 10.1038/sdata.2016.18. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wolf JF, MacKay L, Haworth SE, Cossette M-L, Dedato MN, Young KB, Elliott CI, Oomen RA. Preprinting is positively associated with early career researcher status in ecology and evolution. Ecology and Evolution. 2021; 11 :13624–13632. doi: 10.1002/ece3.8106. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Woolston C. PhDs: the tortuous truth. Nature. 2019; 575 :403–406. doi: 10.1038/d41586-019-03459-7. [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Woolston C. Uncertain prospects for postdoctoral researchers. Nature. 2020; 588 :181–184. doi: 10.1038/d41586-020-03381-3. [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zerhouni EA. NIH in the post-doubling era: Realities and strategies. Science. 2006; 314 :1088–1090. doi: 10.1126/science.1136931. [ PubMed ] [ CrossRef ] [ Google Scholar ]
- eLife. 2023; 12: e78706.
Decision letter
Barbara janssens.
DKFZ (German Cancer Research Center) Heidelberg, Germany
Sarvenaz Sarabipour
Johns Hopkins University Baltimore, United States
Reinhart Reithmeier
University of Toronto Toronto, Canada
In the interests of transparency, eLife publishes the most substantive revision requests and the accompanying author responses.
Decision letter after peer review:
Thank you for submitting your article "Meta-research: The changing career paths of PhDs and postdocs trained at EMBL" to eLife for consideration as a Feature Article. Your article has been reviewed by three peer reviewers, and the evaluation has been overseen by the eLife Features Editor (Peter Rodgers). The three reviewers have agreed to reveal their identity: Barbara Janssens; Sarvenaz Sarabipour; Reinhart Reithmeier.
The reviewers and editors have discussed the reviews and we have drafted this decision letter to help you prepare a revised submission. Please also note that eLife does not permit figures in supplementary materials: instead we allow primary figures to have figure supplements. I can suggest how to accommodate the figures in your supplementary materials if your article is accepted.
The eLife Features Editor will also contact you separately about some editorial issues that you will need to address.
This paper deals with career outcomes for some 2284 PhD graduates and post-doctoral fellows from the European Molecular Biology Laboratories, a prestigious research institute that attracts top talent from around the world – the first of its kind. The paper is rich in data beyond simple career outcomes over time and includes gender, publications and collaboration. The methodology involved internet searches like other studies and was enhanced by robust statistical analyses. This important and timely study fills a gap in our knowledge, highlights the important role that institutions like EMBL play in training the next generation of researchers and innovators, and may stimulate other universities and research institutes to do the same. However, there are a number of points that need to be addressed to make the article suitable for publication.
Essential revisions:
1. It is a great achievement to show career destinations for 89% of the 2284 searched for. However the question is in which cohort the 249 "unknown" belong: it could be more transparent to keep those numbers included also in the detailed cohort analysis. It is also unclear, whether the cohort sizes reflect the actual number of researchers who left EMBL in that time period or whether data were lacking. The cohorts reported here increased about 33% from 2004 to 2012 and another 9% by 2020 (Table 2). Could it be, that for more recent cohorts more data are available – for example due to the fact that younger researchers can be found on social media like LinkedIn?
2. The main and interesting conclusion in the abstract is that of the 45% of alumni not continuing in academic research, one third does industry research and one third is in a science-related profession. Other interesting take-home messages are, that a large proportion of alumni changing sectors enter – or are quickly promoted to – managerial positions, that of those who entered a research position in industry one in ten returned to academia and that female Alumni were found less frequently in PI roles. It would be interesting to know, whether there is a difference in these numbers between cohorts – e.g. if more alumni return from industry to academia in recent cohorts and whether female researchers stay longer in postdoc roles (which could influence the total number of female PIs at a given time point).
3. In the time-resolved analysis the authors claim that the probability of being found as a PI in Academia diminished by about 15% after the first cohort (Figure 2C). However the question is whether the absolute number of PIs also decreased. This could be clarified with some reference numbers in the supplementary materials. When calculating the % of the given cohort sizes at different institutes (supp. Table 4) some differences (increase vs decrease) in the number of PIs can be found. Even if the hypothesis that the differences between cohorts are not EMBL-specific, and reflect a wide-spread change in the number of PhDs and postdocs relative to available PI positions seems valid, it would be good to clarify this issue.
4. Interesting findings is also the significant change in time between PhD and first PI position of 0.8 calendar years between the 2004 and 2012 cohorts. It is not surprising that publication factors are highly correlated with entry into PI positions: indeed all ECRs who became PIs published well, but, not all ECRs who published well became PIs! Publications have become more collaborative over the last decade (the number of coauthors has doubled and the number of first author publications per ECR diminished). Another relevant observation is the lack of correlation with group leader seniority or nationality. Group publications were also predictive for other research and science-related careers. Finally a strong observation is that 45% of leavers from the last 5-years who were found to be working outside of academia held senior or management-level roles. These findings can be reassuring for ECRs and the authors could consider to clearly state these in their conclusions.
5. Regarding the career tracking method used for this study: doing google searches for 2284 Alumni is a plausible effort and has probably been time consuming. A question for other research institutes and universities would be, whether this method of career tracking is scalable and/or feasible to continue as a regular task, or whether the authors see this as a one-time effort. If so, what kind or extent of career tracking would the authors recommend to continue sustainably? Performing career tracking is quite relevant, as institutes worldwide now start to be asked to deliver such data to governments and funding agents.
6. In Discussion and future work, it would be valuable to briefly discuss/aspire that institutions such as EMBL compile and publicly report on this type of data/records analysis together with surveys (what authors call mixed methods) of career intent and research environment.
i. With surveys one could have the number of EMBL trainees that actually applied for PI jobs.
ii. The fact that women were found less frequently in PI jobs does not reveal if (1) women apply less frequently or (2) search committees offer PI jobs less often to women or (3) combination of the two.
iii. Surveys together with the data presented in this work can examine the role of lab environment during training and job application.
7. An observation that authors have in the manuscript is that women are less represented as AcPIs (academic PIs). But it's not possible to claim it's an active mechanism. It would be valuable if authors plot the timeline by gender so readers could see the noise.
8. In various panels of Figures 2-4, please clarify if "Time after EMBL" (the label on the x-axis) means "Time after leaving EMBL" or "Time after arrival at EMBL".
Also, it appears that regardless of the cohort, postdocs have increased chances to become a PI only years after they leave EMBL, why? did they go on to do a second postdoc?
9. Please discuss supplementary table 4 in the text, and highlight any common findings from these studies.
10. The authors may also wish to comment on how some members of faculty recruitment committees may need to be trained to recognize bias in relying too heavily on citation indices and first author publications in hiring decisions rather than the scientific contribution of highly-qualified candidates to collaborative projects.
Barbara Janssens, DKFZ (German Cancer Research Center) Heidelberg, Germany .
Sarvenaz Sarabipour, Johns Hopkins University Baltimore, United States .
Reinhart Reithmeier, University of Toronto Toronto, Canada .
Author response
Essential revisions: 1. It is a great achievement to show career destinations for 89% of the 2284 searched for. However the question is in which cohort the 249 "unknown" belong: it could be more transparent to keep those numbers included also in the detailed cohort analysis. It is also unclear, whether the cohort sizes reflect the actual number of researchers who left EMBL in that time period or whether data were lacking. The cohorts reported here increased about 33% from 2004 to 2012 and another 9% by 2020 (Table 2). Could it be, that for more recent cohorts more data are available – for example due to the fact that younger researchers can be found on social media like LinkedIn?
Thank you for these comments, we are happy to clarify here and in the manuscript:
The cohort sizes reflect the number of all PhD students and postdoctoral fellows who left in that time period, according to the official institutional administrative records. The organization grew during the time period included in the study (1997-2020), and the number of PhD students and postdocs in the later cohorts is therefore greater. It excludes individuals who were marked as deceased in our alumni records at the point the data was originally shared with us (June 2017 for 1997-2016 leavers and April 2021 for 2017-2020 leavers; or whose death we learned of during the update of our career tracking data in summer 2021 (through an updated alumni record, or if we found an online obituary)).
For each PhD or postdoc cohort, the percentage of alumni whose current position is unknown is between 9% and 14%, with no consistent trends with time. However, for the oldest cohort we less frequently found a complete online CV or biosketch that was detailed enough to confirm the type of position held for the full-time span since EMBL, particularly for postdoc alumni. Fewer of the older cohort therefore have a detailed CV/career path and there is a higher percentage of unknowns for specific timepoints after EMBL.
Changes to manuscript:
We have added an additional row with the number and percentage of alumni with detailed career paths for each cohort in Table 2 – new row = n(%) detailed career path
We have also clarified that the increased cohort size is due to growth in sentence that refers to this table: “More recent cohorts were also larger (Table 2), reflecting growth of the organization between 1997 and 2020.”
Column charts showing type of position by cohort (Figure 1B and Figure 1 —figure supplement 2 as 1B, but for other time-points) now include all alumni, not just those with a full career path available.
2. The main and interesting conclusion in the abstract is that of the 45% of alumni not continuing in academic research, one third does industry research and one third is in a science-related profession. Other interesting take-home messages are, that a large proportion of alumni changing sectors enter – or are quickly promoted to – managerial positions, that of those who entered a research position in industry one in ten returned to academia and that female Alumni were found less frequently in PI roles. It would be interesting to know, whether there is a difference in these numbers between cohorts – e.g. if more alumni return from industry to academia in recent cohorts and whether female researchers stay longer in postdoc roles (which could influence the total number of female PIs at a given time point). ‘ if more alumni return from industry to academia in recent cohorts’
This is difficult to assess due to the different career lengths and small numbers transitioning from one type of career to another. For example, of 415 alumni who we have a career path for and had at least one Industry role, just 22 returned to a faculty position. Twelve of these were from the oldest cohort (of 117 who held an industry role), compared to 7 (of 178) for the most recent cohort. Similarly, for PI to industry, 21 transitions were observed from the 539 career paths – 16 from the oldest cohort (from 231) and 1 (from 128) in the newest. Given that the propensity to transition may also change with career length, it is difficult to make comparisons or detect meaningful trends from these small numbers.
“whether female researchers stay longer in postdoc roles (which could influence the total number of female PIs at a given time point).”
We did not observe a statistically significant difference in length between PhD and becoming a PI, but agree that his is interesting and that it should be included in the manuscript.
To add this to the manuscript we made three changes: Additional figure supplement showing the average PhD to PI length for male vs female alumni [as previous figure comparing 2005-2012 and 1997-2004 cohorts, but now comparing female vs male alumni] (Figure 3 —figure supplement 1) – this suggests that male and female researchers spend similar times in postdoc roles as the differences are not statistically significant.
We have now included Kaplan Maier plots by gender, which also illustrate the entry into PI (and other) roles with time (as Figure 3B -C and Figure 3 —figure supplement 2 additional plots for AcOt, IndR, NonSci).
We also expand discussion of these data in the main text – see below with new detail italicised. (note: to allow more detailed discussion without requiring repetition of the information, we have moved this section after the sections on changes in career outcomes, where the Kaplan-Meier and PhD to PI length are first discussed).
Many studies have reported that female ECRs are less likely to remain in an academic career (44, 45). Consistent with these previous studies, we observed that male alumni were found more frequently in PI roles (Figure 3A-B; Table S5 in Supplementary information). Figure 3A-B; Table S5 in Supplementary information. There was no statistically significant difference in the time to obtain a PI role between male and female alumni for alumni from 1997-2012, who became PIs within 9-years (Figure 3—figure supplement 1). The difference in career outcomes is therefore unlikely to be explained by different career dynamics for male and female alumni.
Female alumni were more frequently found in science-related non-research roles than male alumni (Figure 3A). In our Cox models, there was also a statistically significant difference between genders in entry into science-related non-research roles for postdoc alumni [p = 0.016] (Figure 3C; Table S5 in Supplementary information).
We more frequently found male alumni in industry research and non-science-related roles than their female colleagues (Figure 3A; Figure 3—figure supplement 2B-C). However, a higher percentage of female than male alumni could not be located. As academics are usually listed on institutional websites, often with a historical publication list that allows unambiguous identification, we expect that most alumni who were not located are employed in the non-academic sector. This means that, considering the higher percentage of female alumni with unknown career paths (where non-academic careers are likely over-represented), the true percentage of female alumni in industry research and non-science-related roles is likely higher, and possibly comparable with the percentage of male alumni in these roles.
We have added information on the absolute number of PIs for each group in the supplementary table that collates the data published from other institutions and comparable EMBL data (in original manuscript table 4; now Table S3 in Supplementary file 1). We have also expanded discussion of this table the text in response to comment 9 (see comment 9 below) and include the absolute numbers.
Additional clarification
In the datasets, the number of PhDs trained per year has increased with time at all institutions. The cohort size – and how much this has increased for more recent cohorts however varies – for example, Stanford’s 2011-2015 cohort was just 18% larger than its 2000-2005 cohort (503 vs 426), whilst the University of Toronto’s 2012-2015 is 96% larger than its 2000-2003 (1234 vs 629). For EMBL, the PhD cohort sizes increased 45% from 256 for the 1997-2004 cohort, to 372 for 2013-2020. Therefore, the absolute number of PIs with time is difficult to compare between institutions. We therefore feel that the percentage of alumni entering different career options is the most pragmatic measure for comparing how career outcomes are changing with time. It can be viewed as the ‘chance’ of a ECR from a specific programme of entering that career area. If an institution continues to train the same absolute number of PIs per year, but trained many more scientists, it nevertheless saw a big difference the career outcomes of its alumni, with more alumni entering non-PI roles.
We have made the following changes to the manuscript to emphasise the positive aspects of our career findings (but balance the editorial comment that “If possible please shorten the first paragraph of the discussion and avoid any unnecessary repetition of material from earlier in the article.”).
This now reads:
“Many ECRs are employed on fixed-term contracts funded by project-based grants, sometimes for a decade or more (52, 53), and surveys suggest that ECRs are concerned about career progression (18-22). We hope ECRs will be reassured by the results of our time-resolved analysis that indicate that the skills and knowledge developed as an ECR can be applied in academia, industry and other sectors. Within academic research, service and teaching, we observed a marked reduction in the percentage of alumni entering PI roles; nevertheless, academic careers continue to be an important career destination. The percentage of alumni entering career areas outside academic research, service and teaching has increased, and our data suggest that ECRs’ skills are valued in these careers; 45% of leavers from the last 5-years who were found to be working outside of academic research and teaching held senior or management-level roles.”
We have expanded the Discussion section ‘Future career studies’ to include a recommendation as follows:
“Evaluating the outcomes of training programmes, and making these data transparently available is a valuable exercise that can provide information to policymakers, transparency for ECRs, and help institutions provide effective career development support. We plan to update our observational data every four years, and maintain data on the career paths of alumni for 24 years after they leave EMBL. This will help us to identify any further changes in the career landscape and better understand long-term career outcomes. Silva et al. (2019) (73) have also described a method for completing career outcome tracking on a yearly basis with estimations of the time and other resources required. We encourage institutions to consider whether they can adapt our, or Silva’s method to the administrative processes and data-privacy regulations applicable at their institutions.”
6. In Discussion and future work, it would be valuable to briefly discuss/aspire that institutions such as EMBL compile and publicly report on this type of data/records analysis together with surveys (what authors call mixed methods) of career intent and research environment. i. With surveys one could have the number of EMBL trainees that actually applied for PI jobs. ii. The fact that women were found less frequently in PI jobs does not reveal if (1) women apply less frequently or (2) search committees offer PI jobs less often to women or (3) combination of the two. iii. Surveys together with the data presented in this work can examine the role of lab environment during training and job application.
Changes to manuscript: We have re-written the ‘Future career studies’ section of the discussion to incorporate these points. This now reads:
“Future studies should also ideally include mixed-method longitudinal studies. This would allow career motivations, skills development, research environment, job application activity, and other factors to be recorded by surveys during ECR training. Correlating these factors to career and training outcomes would allow investigation of multifactorial and complex issues such as gender differences in career outcomes, and provide policymakers with a fuller picture of workforce trends. Such studies will, however, require large sample sizes from multiple institutions and would need significant time investment and coordination over a long time period. The commitment and the support of funders and institutions would therefore be critical.”
Changes to manuscript: We have generated Kaplan Maier plots by gender for entry into each career area, and include these in Figure 3 and Figure 3 —figure supplement 2 (see discussion of reviewer comment 2, above). The hazard ratios, 95% confidence intervals and p values are provided in a supplementary table in file 1 so that the confidence can be judged.
We have added a clarification in the figure legend (“Time after EMBL refers to the number of calendar years between defence of an EMBL PhD and first PI role (for PhD alumni)) – or number of calendar years between leaving the EMBL postdoc programme and first PI role (for postdoc alumni)”, or clarified this in the figure labels, for each figure.
We have expanded on this in the Results section ‘EMBL alumni contribute to research and innovation in academic and non-academic roles’, adding the following text:
“On average, PhD alumni became PIs 6.1 calendar years after their PhD defence. For postdoc alumni, the start year of the first PI role was on average 7.3 calendar years after their PhD and 2.5 years after completing their EMBL postdoc. Almost half of EMBL postdoc alumni who became PIs did so directly after completing their EMBL postdoc (168 of 343 alumni with a detailed career path available). Other postdoc alumni made the transition later, most frequently after one additional postdoc (71 alumni) or a single academic research / teaching / service position (56 alumni). Some alumni held multiple academic (40 alumni), or one or more non-academic positions (8 alumni) between their EMBL postdoc and first PI role.”
And to provide balance / to avoid focusing only on academic careers, we also include an additional sentence in the subsequent paragraph on non-academic areas:
“The average time between being awarded a PhD and the first industry research, science-related or non-science-related role was 5.0, 5.3 and 4.2 calendar years respectively.”
We have expanded the discussion of this in the Results section ‘The percentage of EMBL alumni becoming PIs is similar to data released by North American institutions for both older and more recent cohorts’. [note that due to rearrangements, supplementary table 4 is now ‘Table S3 in Supplementary file 1’]. This now reads:
“A number of institutions have released cohort-based PhD outcomes data online or in publications (32, 44-49). Of these, a recent dataset from Stanford University offers the longest career tracking, reporting outcomes for PhD graduates from 20 graduation years (2000-2019) (44). In this dataset, Stanford University reported that 34% (145/426) of its 2000-2005 PhD alumni were in research-focussed faculty roles in 2018. This is comparable to the 37% (78/210) we observe for the EMBL alumni for the same time period. For 2011-2015 graduates, comparable percentages of Stanford (13%, 63/503) and EMBL (11%, 25/234) graduates were also in PI roles in 2018 (Figure 1C). Figure 1 – Supplement 3 plots data from five other datasets alongside equivalent data from EMBL (further details of each dataset are available in the Table S3 in Supplementary file 1). This includes a published dataset from the University of Toronto (32, 49), which reported that 31% (192/629) of its 2000-2003 life science division graduates and 25% (203/816) of its 2004-2007 life science graduates were in tenure stream roles in 2016. The equivalent EMBL data is comparable at 39% (52/132) and 28% (49/172) of graduates in PI roles respectively. Across all six datasets, EMBL and the other institutes generally have a similar proportion of alumni entering PI roles for comparable cohorts. This is consistent with our hypothesis that the differences between cohorts are not EMBL-specific, and reflect a wide-spread change in the number of PhDs and postdocs relative to available PI positions.”
We have expanded this section of the discussion (third paragraph of ‘Addressing ECR career challenges’ in the discussion), also mentioning the trend to narrative CVs that has accelerated recently. This now reads:
“Research assessment and availability of funding play an important role in determining the career prospects of an academic. Therefore, it is also vital that factors that may lead to misperception of the productivity of ECRs, such as involvement in large-scale projects, career breaks, or time spent on teaching and service, are considered in research assessment during hiring, promotion and funding decisions. Initiatives such as the San Francisco Declaration on Research Assessment (DORA) and Coalition for Advancing Research Assessment (CoARA) have been advocating for an increased focus on good practice in evaluating research outputs, and many institutions and funders have reviewed their practices. Cancer Research UK, for example, now asks applicants to its grants to describe three to five research achievements, which can include non-publication outputs (64) and narrative CV formats that allow candidates to put their achievements in context are also becoming more common (61). The impact of the coronavirus pandemic on research productivity of researchers with caregiving responsibilities makes such actions imperative (65-67). We welcome this increased focus on qualitative assessment of scientific contribution, rather than reliance on publication metrics. To ensure successful implementation and to avoid unintended consequences (such as introducing new biases), it will be important for funders and institutions to provide appropriate guidance and/or training to evaluators and to carefully monitor implementation. Other initiatives that may help ECR involved in ambitious projects to demonstrate their contributions include more transparent author contribution information in publications (68, 69) and promotion of "FAIR" principles of data management (70). The increasing use of preprints (41, 71) is also likely to have a positive effect on the careers of ECRs involved in projects with longer time scales (72).”
Applications invited for the 2024 FIMM-EMBL International PhD Programme
As part of the Nordic EMBL Partnership for Molecular Medicine , we welcome applications for the 2024 FIMM-EMBL International PhD Program. Detailed information regarding the position descriptions, research areas, qualifications, and application and selection process can be found at the University of Helsinki Open Positions page.
We currently have two to four openings for doctoral researchers who show outstanding potential to become independent researchers and who will significantly benefit from the FIMM-EMBL international and interdisciplinary PhD rotational training. Specifically, these positions are in the fields of artificial intelligence, bioinformatics, biomedicine, bioscience, computational neuroscience, drug research, genomics, molecular data science, and molecular medicine .
Outstanding PhD researchers will be recruited to the rotation program where they will work with 2-3 different research groups during a 6-9 month period before selecting a research group in which to remain for their complete PhD studies. This program provides an exciting opportunity to explore and combine different research areas and working environments as well as to learn a variety of science and technologies, making connections within and beyond the institute.
Doctoral researchers in the Rotation Program:
- Gain formal research experience before the doctoral program period
- Diversify and deepen scientific knowledge
- Develop and refine scientific interests
- Receive early-career training with feedback in scientific communication
- Foster a best match with a research mentor(s)
Fully Funded PhD Positions in Life Sciences at EMBL

Would you like to contribute your creativity to an international team of scientists from various disciplines focusing on basic research in the area of molecular life sciences?
Here is an offer from the EMBL International PhD Programme – 2023 Summer Recruitment:
The European Molecular Biology Laboratory (EMBL) invites you to apply for PhD positions in Heidelberg, Barcelona, Grenoble, Hamburg, Hinxton (near Cambridge) and Rome. The EMBL International PhD Programme (EIPP) provides comprehensive interdisciplinary training, maintaining a careful balance between theory and practice, close mentoring and creative freedom, collaborative teamwork and independence. These key characteristics make EIPP a role model, which has inspired similar programmes at research institutions throughout Europe and the World.
Find out more about the cutting-edge Research Topics investigated across our different Research Units .
We welcome candidates with diverse backgrounds, such as in Biology, Chemistry, Physics, Mathematics, Computer Science, Engineering and Molecular Medicine. Applicants must hold, or anticipate receiving before starting at EMBL, a university degree that would formally qualify them to enter a PhD or equivalent programme in the country where the degree was obtained.
Why join us
EMBL provides PhD students with a starting platform for a successful career in science by fostering early independence and interdisciplinary research. The enriching encounter of different nationalities, the friendly and collaborative atmosphere, and the passion for science is what unites EMBL’s diverse staff and provides an ideal setting to forge long-lasting connections and make studying at EMBL a formative experience. Our PhD positions are fully funded and offer broad health care and pension benefits.
Full details on how to submit your online application are available here .
The deadline for submitting the application is 3 April 2023 . Recruited candidates would start their work at EMBL latest by mid of October 2023.
EMBL is a signatory of DORA. Find out how we implement best practices in research assessment in our recruitment processes here .
For further information, please visit our web page or contact the EMBL Graduate Office .
EMBL Australia
A commitment to leading australian research.
EMBL Australia creates opportunities to internationalise Australian research, empowering and training our best early-career researchers and research leaders.
Our unique model offers the best young research leaders up to nine years of security, enabling them to take risks and ask big questions.
- Research Groups
- Scientific Leadership
Providing students with global opportunities
EMBL Australia gives the nation’s best PhD students the opportunity to develop international networks and alliances in Europe via EMBL’s programs, workshops and conferences.
We also provide training programs for PhD students in Australia, giving them a head start in their science careers.
- Travel Grants
- Postgraduate Symposium
- EMBL Australia PhDs
Discovery to plug wound-healing's billion dollar drain on health system
News / 27 March 2024
Read More icon-cheron-right-circled
EMBL Australia Newsletter - March 2024
Newsletters / 26 March 2024
Annette Wittmann joins EMBL Australia as COO
News / 5 March 2024
Cracking the mystery of how HIV gets into the cell’s centre
News / 25 January 2024
Freeze, Frame, Focus: Mastering Cryo-Electron Tomography Techniques
27/05/2024 to 31/05/2024
Future-proofing Computational Proteomics Workshop
Quantitative microscopy for cellular dynamics symposium, creative careers in science: it all started in the lab.
30/11/2023 to 30/11/2023
Vaishnavi Ananthanarayanan
Senthil arumugam, michelle boyle.
For access to cutting-edge infrastructure, visit EMBL’s Scientific Services
🗞️Our latest newsletter has landed! Attend our #CryoEM training course, cracking the mystery of how #HIV gets into the cell's centre, #lightmicroscopy in focus & much more! Read now: https://bit.ly/3TQtPFp
Subscribe to the EMBL Australia Newsletter
Keep up to date with all the latest EMBL Australia news and events.

- Feature Article
Meta-Research: The changing career paths of PhDs and postdocs trained at EMBL
- Britta Velten
- Bernd Klaus
- Mauricio Ramm
- Wolfgang Huber
- Genome Biology Unit, European Molecular Biology Laboratory, Germany ;
- EMBL International Centre for Advanced Training, European Molecular Biology Laboratory, Germany ;
- Open access
- Copyright information
- Comment Open annotations (there are currently 0 annotations on this page).
- 8,427 views
- 1 citations
Share this article
Cite this article.
- Rachel Coulthard-Graf
- Copy to clipboard
- Download BibTeX
- Download .RIS
- Figures and data
Introduction
Statistical model, data availability, decision letter, author response, article and author information.
Individuals with PhDs and postdoctoral experience in the life sciences can pursue a variety of career paths. Many PhD students and postdocs aspire to a permanent research position at a university or research institute, but competition for such positions has increased. Here, we report a time-resolved analysis of the career paths of 2284 researchers who completed a PhD or a postdoc at the European Molecular Biology Laboratory (EMBL) between 1997 and 2020. The most prevalent career outcome was Academia: Principal Investigator (636/2284=27.8% of alumni), followed by Academia: Other (16.8%), Science-related Non-research (15.3%), Industry Research (14.5%), Academia: Postdoc (10.7%) and Non-science-related (4%); we were unable to determine the career path of the remaining 10.9% of alumni. While positions in Academia (Principal Investigator, Postdoc and Other) remained the most common destination for more recent alumni, entry into Science-related Non-research, Industry Research and Non-science-related positions has increased over time, and entry into Academia: Principal Investigator positions has decreased. Our analysis also reveals information on a number of factors – including publication records – that correlate with the career paths followed by researchers.
Career paths in the life sciences have changed dramatically in recent decades, partly because the number of early-career researchers seeking permanent research positions has continued to significantly exceed the number of positions available ( Cyranoski et al., 2011 ; Schillebeeckx et al., 2013 ). Other changes have included efforts to improve research culture, growing concerns about mental health ( Evans et al., 2018 ; Levecque et al., 2017 ), increased collaboration ( Vermeulen et al., 2013 ), an increased proportion of project-based funding ( Lepori et al., 2007 ; Jonkers and Zacharewicz, 2016 ) and greater awareness of careers outside academic research ( Hayter and Parker, 2019 ). Nevertheless, many PhD students and postdocs remain keen to pursue careers in research and, if possible, secure a permanent position as a Principal Investigator (PI) at a university or research institute ( Fuhrmann et al., 2011 ; Gibbs et al., 2015 ; Lambert et al., 2020 ; Roach and Sauermann, 2017 ; Sauermann and Roach, 2012 ).
Data on career paths in the life sciences have become increasingly available in recent years ( Blank et al., 2017 ; Council for Doctoral Education, 2020 ), and such data are useful to individuals as they plan their careers, and also to funding agencies and institutions as they plan for the future. In this article we report the results of a time-resolved analysis of the career paths of 2284 researchers who completed a PhD or postdoc at the European Molecular Biology Laboratory ( EMBL ) between 1997 and 2020. This period included major global events, such as financial crisis of 2007 and 2008 ( Izsak et al., 2013 ; Pellens et al., 2018 ), and also major events within the life sciences (such as the budget of the US National Institutes of Health doubling between 1998 and 2003 and then plateauing; Wadman, 2012 ; Zerhouni, 2006 ).
EMBL is an intergovernmental organisation with six sites in Europe, and its missions include scientific training, basic research in the life sciences, and the development and provision of a range of scientific services. The organization currently employs more than 1110 scientists, including over 200 PhD students, 240 postdoctoral fellows, and 80 PIs. EMBL has a long history of training PhD students and postdocs, and the EMBL International PhD Programme – one of the first structured PhD programmes in Europe – has a completion rate of 92%, with students taking an average of 3.95 years to submit their thesis (data for 2015–2019). More recently, EMBL has launched dedicated fellowship programmes with structured training curricula for postdocs.
Data collection for this study was initially carried out in 2017 and updated in 2021. Using manual Google searches, we located publicly available information identifying the current role of 89% (2035/2284) of the sample ( Table 1 ). These alumni were predominantly based in the European Union (60%, 1224/2035), other European countries including UK and Switzerland (20%), and the US (11%). For 71% of alumni (1626/2284), we were able to reconstruct a detailed career path based on online CVs and biographies (see Methods). EMBL alumni also ended up in a range of careers, which were classified as follows: Academia: Principal Investigator; Academia: Postdoc; Academia: Other research/teaching/service role; Industry Research; Science-related Non-research; and Non-science-related. We also collected data on different types of jobs within the last three of these career areas ( Table 2 ).
Career outcomes for 2284 EMBL alumni.
See Table 2 for more information on the different jobs covered by Industry Research, Science-related Non-research, and Non-science-related. This classification is based on Stayart et al., 2020 .
AcPI: includes those leading an academic research team with financial and scientific independence – evidenced by a job title such as group leader, professor, associate professor or tenure-track assistant professor. Where the status was unclear from the job title, we classified an alumnus as a Principal Investigator (PI) if one of the following criteria was fulfilled: (a) they appear to directly supervise students/postdocs (based on hierarchy shown on website); (b) they have published a last author publication from their current position; (c) their group website or CV indicates that they have a grant (not just a personal merit fellowship) as a principal investigator. AcOt: differs from Stayart et al., 2020 in that it includes academic research, scientific services or teaching staff (e.g., research staff, teaching faculty and staff, technical directors, research infrastructure engineers).
Classification of Industry Research, Science-related Non-research and Non-science-related positions.
This function differs from the schema in Stayart et al., 2020 ; it includes alumni carrying out or overseeing scientific research in industry as group leaders, research staff, technical directors and non-directorship research leadership roles, including alumni who appear to be working in computational biology roles of a pharma, biotech, contract research or similar company regardless of job title (i.e. including data science roles that appear to be related to analysis of research-related data.)
Founders of companies whose primary focus is R&D (including contract research organizations).
Includes director-level senior management roles overseeing the scientific direction & research of a company with R&D focus, e.g. CSOs in biotech start-ups.
Not including computational biology roles linked to R&D functions.
On average, the alumni in our sample published an average of 4.5 research articles about their work at EMBL, and were the first author on an average of 1.6 of those articles (Table S1 in Supplementary file 1 ). Overall, 90% of the sample (2047/2284) authored at least one article about their EMBL work, and 73% (1666/2284) were the first author on at least one article. The average time between being awarded a PhD and taking up a first role in a specific career area ranged from 4.2 years for Non-science-related positions to 6.8 years for a Principal Investigator (PI) position.
Most alumni remain in science
The majority of alumni (1263/2284=55.3%) were found to be working in an academic position in 2021, including 636 who were PIs, 244 who were in Academia: Postdoc positions, and 383 who were working in Academia: Other positions, which included teaching, research and working for a core facility/technology platform ( Figure 1A ). Just under one-sixth (332/2284=14.5%) were employed in Industry Research positons, and a similar proportion (349/2284=15.3%) were employed in Science-related Non-research positions, such as technology transfer, science administration and education, and corporate roles at life sciences companies. Around 4% were employed in professions not related to science, and the current careers of around 11% of alumni were unknown.
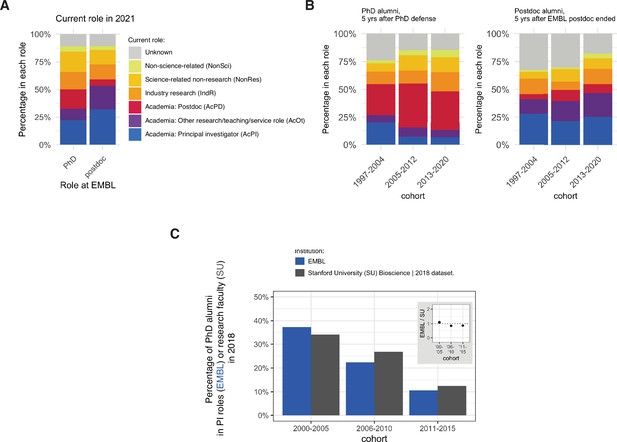
Career outcomes for EMBL alumni.
( A ) Charts showing the percentage of PhD alumni (n=969) and postdoc alumni (n=1315) from EMBL in different careers in 2021 (see Table 1 ). ( B ) Charts showing percentage of PhD (left, n=800) and postdoc (right, n=1053) alumni in different careers five years after finishing their PhD or postdoc, for three different cohorts. Chart excludes 169 PhD students and 262 postdocs who have not yet reached the five-year time point. ( C ) Charts showing the percentage of PhD alumni from EMBL (blue column) in PI positions with the percentage of PhD alumni from Stanford University (grey column) in research-focused faculty positions ( Stanford Biosciences, 2021 ). Detailed information about the comparison group can be found in Table S3 in Supplementary file 1 .
Figure 1—source data 1
Summary data plotted in Figure 1A, B .
Of those who became PIs, 75.3% moved from a postdoc to their first PI position, with 20.6% moving from an Academia: Other position ( Figure 1—figure supplement 1A ). On average, PhD alumni became PIs 6.1 calendar years after their PhD defence, and postdoc alumni became PIs 2.5 years after completing their EMBL postdoc. Almost half of the postdoc alumni who became PIs did so directly after completing their EMBL postdoc (168 of 343). Other postdoc alumni made the transition later, most frequently after one additional postdoc (71 alumni) or a single Academia: Other position (56 alumni). 40 alumni held multiple academic positions between their EMBL postdoc and their first PI position, and eight had one or more non-academic positions during this period.
The career paths of those in other positions were more varied ( Figure 1—figure supplement 1B–E ). For example, for alumni who moved into Industry Research, 20.2% entered their first industry role directly from their PhD, 56.4% from a postdoc position, and 13.3% from Academia: Other positions. Moreover, 71.6% remained in this type of role long-term.
The wide variation in job titles used outside academia makes it difficult to assess career progression, but almost 60% (453/766) of alumni working outside academia had a current job title that included a term indicative of a management-level role (such as manager, leader, senior, head, principal, director, president or chief). For leavers from the last five years (2016–2020), this number was 45% (78/174), suggesting that a large proportion of the alumni who leave academia enter – or are quickly promoted to – managerial positions.
For further analysis, EMBL alumni were split into three 8 year cohorts. More recent cohorts were larger, reflecting the growth of the organization between 1997 and 2020, and also contained a higher percentage of female researchers ( Table 3 ). When comparing cohorts, we observed some differences in the specific jobs being done by alumni outside academia 2021 (Table S2 in Supplementary file 1 ). For example, the percentage of alumni involved in ‘data science, analytics, software engineering’ roles increased from 2% (11/625) for the 1997–2004 cohort to 4% (37/896) for the 2013–2020 cohort. However, the absolute number of alumni for most jobs outside academia was small, so our time-resolved analysis therefore focussed on the broader career areas described above.
Overview of PhD and postdoc cohorts.
Percentage of embl alumni who become pis is similar to that for other institutions.
For all timepoints, the percentages of alumni from the 2005–2012 and 2013–2020 cohorts working in PI positions in 2021 were lower than the percentage for the 1997–2004 cohort ( Figure 1—figure supplement 2 ). To assess whether this pattern was specific to EMBL, we compared our data with data from other institutions, noting that different institutions can use different methods to collect data and classify career outcomes. We also note that career outcomes are influenced by the broader scientific ecosystem and by the subject focus of institutions and departments, which may attract early-career researchers with dissimilar career motivations. Nevertheless, comparing long-term outcomes with other institutions allows us to interrogate whether the changes we observe for the most frequent, well-defined and linear career path – the PhD–>Postdoc–>PI career path – reflect a general trend.
A number of institutions have released data on career outcomes for PhD students. Stanford University, for example, has published data on the careers of researchers who received a PhD between 2000 and 2019 ( Stanford Biosciences, 2021 ): Stanford has reported that 34% (145/426) of its 2000–2005 PhD alumni were in research-focussed faculty roles in 2018, and that 13% (63/503) of its 2011–2015 PhD alumni were in PI roles; these numbers are comparable to the figures of 37% (78/210) and 11% (25/234) we observe for EMBL alumni for the same time periods ( Figure 1C ). The EMBL data are also comparable to data from the life science division at the University of Toronto ( Reithmeier et al., 2019 ; University Toronto, 2021 ): for example, Toronto has reported that 31% (192/629) of its 2000–2003 graduates and 25% (203/816) of its 2004–2007 graduates were in tenure stream roles in 2016; the corresponding figures for EMBL were 39% (52/132) and 28% (49/172).
We also compared our EMBL data with data from the University of Michigan, the University of California at San Francisco, and the University of Chicago, and found similar proportions of alumni entering PI positions for comparable cohorts ( Figure 1—figure supplement 3 ). This is consistent with our hypothesis that the differences between cohorts are not EMBL-specific, and reflect a wide-spread change in the number of PhDs and postdocs relative to available PI positions.
We did not analyse the data for other career outcomes, as the smaller numbers of individuals in these careers made it difficult to identify real trends. Moreover, only a small number of institutions have released detailed data on the career destinations of recent postdoc alumni, and we are not aware of any long-term cohort-based data.
The proportion of EMBL alumni who become PIs has decreased with time
To estimate the probability of alumni from different cohorts entering a specific career each year after completing a PhD or postdoc at EMBL, we fitted the data to a Cox proportional hazards model. This is a statistical regression method that is commonly used to model time-to-event distributions from observational data with censoring (i.e., when not all study subjects are monitored until the event occurs, or the event never occurs for some of the subjects). In brief, we fitted the data to a univariate Cox proportional hazards model to calculate hazard ratios, which represent the relative chance of the event considered (here: entering a specific career) occurring in each cohort with respect to the oldest cohort. We also calculated Kaplan–Meier estimators, which estimate the probability of the event (entering a specific career) at different timepoints.
For both PhD and postdoc alumni entering PI positions, we observe hazard ratios of less than one in the Cox models when comparing the newer cohorts with the oldest cohort (Table S4 in Supplementary file 1 ), which indicates that the chances of becoming a PI have become lower for the newer cohorts. The Kaplan–Meier plots illustrate lower percentages of PIs among alumni from the most recent cohorts compared to the oldest cohort at equivalent timepoints ( Figure 2A ). Nevertheless, becoming a PI remained the most common career path for alumni from the 2005–2012 cohort (90/341=26.4% for PhD alumni) and (123/422=29.1% for postdoc alumni), and the most recent cohort of alumni appear to be on a similar trajectory.
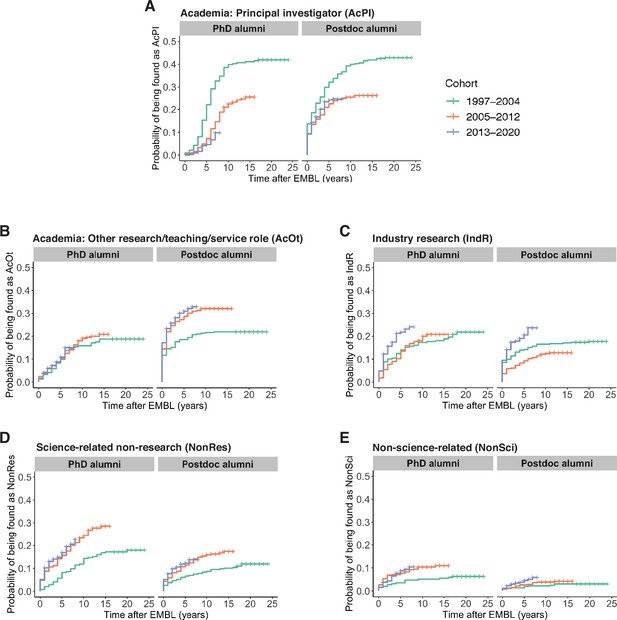
Changes in career outcomes for more recent cohorts.
( A ) Kaplan–Meier plots showing the estimated probability of an individual being in a PI position (y-axis) as a function of time after EMBL (x-axis) for three cohorts of PhD alumni (left) and three cohorts of postdoc alumni (right). Time after EMBL refers to the number of calendar years between PhD defence or leaving the EMBL postdoc programme and first PI position. ( B–E ) Similar Kaplan–Meier plots for Academia: Other positions ( B ), Industry Research positions ( C ), Science-related Non-research positions ( D ), and Non-science-related professions ( E ). Hazard ratios calculated by a Cox regression model can be found in Table S4 in Supplementary file 1 .
Kaplan–Meier plots show increased proportions of the 2005–2012 and 2013–2020 cohorts entering Science-related Non-research and Non-science-related positions, compared to the 1997–2004 cohort for both PhD and postdoc alumni ( Figure 2D, E ). For the most recent (2013–2020) cohort, there was also an increased rate of entry into Industry: Research positions compared to alumni from PhD and postdoc cohorts from 1997 to 2004 and 2005–2012 ( Figure 2C , Table S4 in Supplementary file 1 ). For Academia: other positions, the rate of entry was similar for all three PhD cohorts, though some differences between cohorts were observed for postdoc alumni ( Figure 2B ).
A small increase in time between year of PhD and first PI position
We decided to explore to what extent increasing postdoc length may contribute to the decreased proportion of alumni who are found as PIs in the years after leaving EMBL. In order to fairly compare alumni from different cohorts, we included only alumni for whom we had a detailed career path, who had defended their PhD at least nine years ago, and who had become a PI within nine years of defending their PhD. We chose a nine-year cut-off because this was the time interval between the last PhDs in the 2005–2012 cohort and the execution of this study; moreover, for PhD alumni from the oldest cohort (1997–2004), most of those who became PIs had done so within nine years (89/97=92%).
157 of the PhD alumni in our sample met these criteria, taking an average of 5.6 calendar years to become a PI (see Methods). There was a statistically significant difference in the average time from PhD to first PI position between the 1997–2004 cohort (5.2 years) and the 2005–2012 cohort (6.1 years; Figure 2—figure supplement 1A ). 218 of the postdoc alumni in our sample met these criteria, taking an average of 2.5 calendar years to become a PI after leaving EMBL (see Methods). There was no statistically significant difference in time between EMBL and first PI role for the 1997–2004 and 2005–2012 postdoc cohorts ( Figure 2—figure supplement 1B ). However, the time between receiving their PhD and becoming a PI increased by from 5.3 calendar years for the 1997–2004 postdoc cohort to 6.0 calendar years for the 2005–2012 postdoc cohort ( Figure 2—figure supplement 1C ).
Gender differences in career outcomes
Many studies have reported that female early-career researchers are less likely to remain in academia ( Alper, 1993 ; Martinez et al., 2007 ). Consistent with these studies, male alumni from EMBL were more likely than female alumni to end up in a PI position ( Figure 3A and B ; Table S5 in Supplementary file 1 ). However, for alumni from 1997 to 2012, there was no statistically significant difference in the length of time taken by male and female alumni to become PIs ( Figure 3—figure supplement 1 ). Female alumni were more likely to end up in a Science-related Non-research position, and male alumni were more likely to end up in an Industry Research or Non-science-related position ( Figure 3 ; Figure 3—figure supplement 2 ). However, female alumni were also more likely to be classified as unknown, and since it is more difficult to follow careers outside the academic world, it is possible that the number of women who established careers outside academia (in positions such as Industry Research, Science-related Non-research, and Non-science-related) is higher than our results suggest.
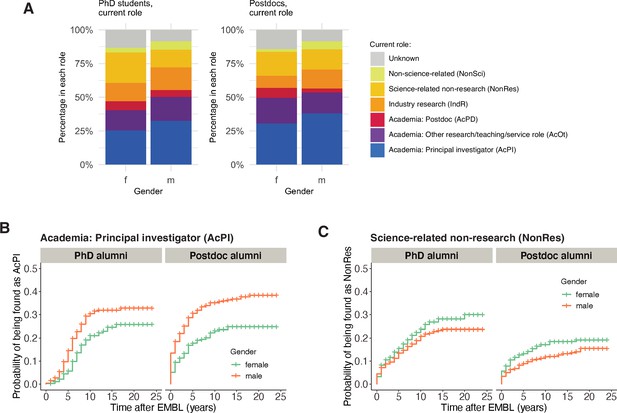
Gender differences in career outcomes.
( A ) Charts showing the percentage of female (n=415) and male (n=554) PhD alumni, and female (n=492) and male (n=823) postdoc alumni, in different careers in 2021. ( B ) Kaplan–Meier plots showing the estimated probability of an individual being in a PI position (y-axis) as a function of time after EMBL (x-axis), stratified by gender for PhD alumni (left) and postdoc alumni (right). ( C ) Kaplan–Meier plots showing the estimated probability of an individual being in a science-related non-research position as a function of time after EMBL, stratified by gender for PhD alumni (left) and postdoc alumni (right). Kaplan–Meier plots for other career outcomes are shown in Figure 3—figure supplement 2 . Hazard ratios calculated by a Cox regression model can be found in Table S5 in Supplementary file 1 .
Figure 3—source data 1
Summary data plotted in Figure 3A .
Future PIs, on average, published more papers while at EMBL
Publication metrics have been linked to the likelihood of obtaining ( van Dijk et al., 2014 ; Tregellas et al., 2018 ) and succeeding ( von Bartheld et al., 2015 ) in a faculty position. In this study, alumni who became PIs had more favourable publication metrics from their EMBL work – for example, they published more articles, and their papers had higher CNCI values. (CNCI is short for Category Normalized Citation Impact, and a CNCI value of one means that the number of citations received was the same as the average for other articles in that field published in the same year; Figure 4A and B ; Table S6 in Supplementary file 1 ). Using univariate Cox models for time to PI as a function of number of first-author research articles from EMBL work, we estimated that a postdoc with one first-author publication was 3.2 times more likely to be found in a PI position than a postdoc without a first-author publication (95% confidence interval [2.2, 4.7]), and a post-doc with two or more first-author publications was 6.6 times more likely (95% confidence interval [4.7, 9.3]; Figure 4C ).
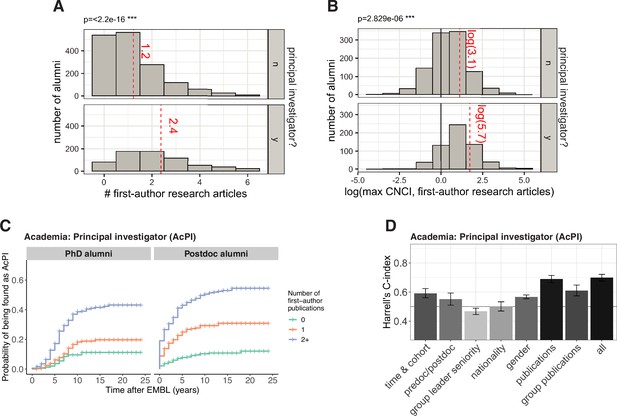
Publication factors are highly correlated with becoming a PI.
( A ) Histograms showing the number of alumni who have 0, 1, 2, 3,... first-author articles from their time at EMBL and became PIs (bottom; n=662, excluding 23 outliers), and did not become PIs (top; n=1594, excluding 5 outliers). For clearer visualization, and to protect the identity of alumni with outlying numbers of publications, the x-axis is truncated at the 97.5 th percentile. The mean for each group (including outliers) is shown as a red dashed line; alumni who became PIs have an average of 2.4 first-author articles from their time at EMBL, whereas other alumni have an average of 1.2 articles; this difference is significant ( P <2.2 × 10 –16 ; Welch’s t-test). ( B ) 1656 alumni had one or more first-author articles from their time at EMBL that had a CNCI value in the InCites database. For each of these alumni, the natural logarithm of the highest CNCI value was calculated, and these histograms show the number of alumni for which this natural logarithm is between –4.5 and –3.5, between –3.5 and –2.5, and so on; the bottom histogram is for alumni who became PIs, and the top histogram is for other alumni. A CNCI value of 1 (plotted here at ln(1)=0; vertical black line) means that the number of citations received by the article is the same as the average for other articles in that field published in the same year. The mean for each group is shown as a red dashed line; alumni who became PIs have an average CNCI of 5.7, whereas other alumni have an average CNCI of 3.1; this difference is significant ( P <2.829 × 10 –6 ; Welch’s t-test). ( C ) Kaplan–Meier plots showing the estimated probability of an individual becoming a PI (y-axis) as a function of time after EMBL (x-axis), stratified by number of first-author publications from research completed at EMBL, for PhD alumni (left) and postdoc alumni (right). Hazard ratios calculated by a Cox regression model can be found in Table S7 in Supplementary file 1 . ( D ) Harrell’s C-Index for various Cox models for predicting entry into PI positions. The first seven bars show the C-index for univariate and multivariate models for a subset of covariates (which subset is shown below the x-axis), and the eighth bar is for a multivariate model that includes the covariates from all subsets. The subsets are time & cohort (multivariate, including the variables: cohort, PhD year (if known), start and end year at EMBL), predoc (ie PhD student)/postdoc (univariate), group leader seniority (univariate), nationality (univariate), gender (univariate), publications (multivariate: containing variables related to the alumni’s publications from their EMBL work; these are variables with a name beginning with “pubs” in Table S1 in Supplementary file 1 ) and group publications (multivariate: containing variables related to the aggregated publication statistics for all PhD students and postdocs who were trained in the same group; these are variables with a name beginning with “group_pubs” in Table S1 in Supplementary file 1 ). A value of above 0.5 indicates that a model has predictive power, with a value of 1.0 indicating complete concordance between predicted and observed order to outcome (e.g. entry into a PI position). Bars denote the mean, and the error bars show the 95% confidence intervals. A value of above 0.5 indicates that a model has predictive power, with a value of 1.0 indicating complete concordance between predicted and observed order to outcome (e.g. entry into a PI position). Bars denote the mean, and the error bars show the 95% confidence intervals.
Figure 4—source data 1
Summary data plotted in Figure 4A, B and D .
Publication factors are highly predictive of entry into a PI position
To understand the potential contribution of publication record in the context of other factors – including cohort, gender, nationality, publications, and seniority of the supervising PI – we fitted multivariate Cox models. To quantify publication record, we considered a range of metrics including journal impact factor, which has been shown to statistically correlate with becoming a PI in some studies ( van Dijk et al., 2014 ) and has been used by some institutions in research evaluation ( McKiernan et al., 2019 ). It should be stressed, however, that EMBL does not use journal impact factors in hiring or evaluation decisions, and is a signatory of the San Francisco Declaration on Research Assessment (DORA) and a member of the Coalition for Advancing Research Assessment (CoARA).
To evaluate the predictive power of each Cox model, we used the cross-validated Harrell’s C-index, which measures predictive power as the average agreement across all pairs of individuals between observed and predicted temporal order of the outcome (in our case, entering a specific type of position; see Methods). A C-index of 1 indicates complete concordance between observed and predicted order. For example, for a model of entry into PI roles, a C-index of 1 would mean that the model correctly predicts, for all pairs of individuals, which individual becomes a PI first based on the factors included in the model. A C-index 0.5 is the baseline that corresponds to random guessing. Prediction is clearly limited by the fact that we could not explicitly encode some covariates that are certain to play an important role in career outcomes, such as career preferences and relevant skills. Nevertheless, the C-index for models containing all data were between 0.61 (entry to Industry Research, Figure 4—figure supplement 1B ) and 0.70 (entry into PI positions, Figure 4D ), suggesting that the factors have some predictive power.
To investigate which factors were most predictive for entry into different careers, we compared models containing different sets of factors. Consistent with previous studies, we found that statistics related to publications were highly predictive for entry into a PI position: a multivariate model containing only the publication statistics performs almost as well as the complete multivariate model, reaching a C-index of 0.69 ( Figure 4D ). The publications of the research group the alumnus was trained in (judged by the aggregated publication statistics for all PhD students and postdocs who were trained in the same group) was also predictive, with a C-index of 0.61.
Cohort/year, gender, and status at EMBL (PhD or postdoc) were also predictors of entry into a PI position in our Cox models, with C-indexes of 0.59, 0.57 and 0.55, respectively ( Figure 4D ). This is consistent with our observation that alumni from earlier cohorts/years ( Figure 1B ), male alumni ( Figure 3A ) and postdoc alumni ( Figure 1A ) were more frequently found in PI positions. Models containing only nationality or group leader seniority were not predictive.
For Academia: Other positions, the factors that were most predictive were those related to publications of the research group the alumnus was trained in ( Figure 4—figure supplement 1A ). It is unclear why this might be, but we speculate that this could reflect publication characteristics specific to certain fields that have a high number of staff positions, or other factors such as the scientific reputation, breadth or collaborative nature of the research group and its supervisor. The group’s publications were also predictive for Industry Research and Science-related Non-research positions.
Time-related factors (i.e., cohort, PhD award year and EMBL contract start/end years) were the strongest prediction factors for Industry Research, Science-related Non-research, and Non-science-related positions ( Figure 4—figure supplement 1B–D ), and more recent alumni were more frequently found in these careers ( Figure 2C–E ).
Overall, statistics related to an individual’s own publications were a weak predictor for entry into positions other than being a PI ( Figure 4—figure supplement 1 ; Figure 4—figure supplement 2 ; Table S7–S11 in Supplementary file 1 ). For example, for Industry Research, a model containing statistics for an individual’s publications had a C-index of only 0.53, compared to 0.61 for the complete model, and there were no differences in likelihood of a PhD alumnus with 0, 1 or 2+publications entering an Industry Research position.
Changes in the publications landscape
Reports suggest that the number of authors on a typical research article in biology has increased over time, as has the amount of data in a typical article ( Vale, 2015 ; Fanelli and Larivière, 2016 ); a corresponding decrease in the number of first-author research articles per early-career researcher has also been reported ( Kendal et al., 2022 ). For articles linked to the PhD students and postdocs in this study, the mean number of authors per article has more than doubled between 1995 and 2020 ( Figure 5A ). The mean number of articles per researcher did not change between the three cohorts studied ( Figure 5B ; the mean was 3.6 articles per researcher), but researchers from the second and third cohorts published fewer first-author articles than those from the first cohorts ( Figure 5C ). However, more recent articles had higher CNCI values ( Figure 5D ). The proportion of EMBL articles that included international collaborators also increased from 47% in 1995 to 79% in 2020.
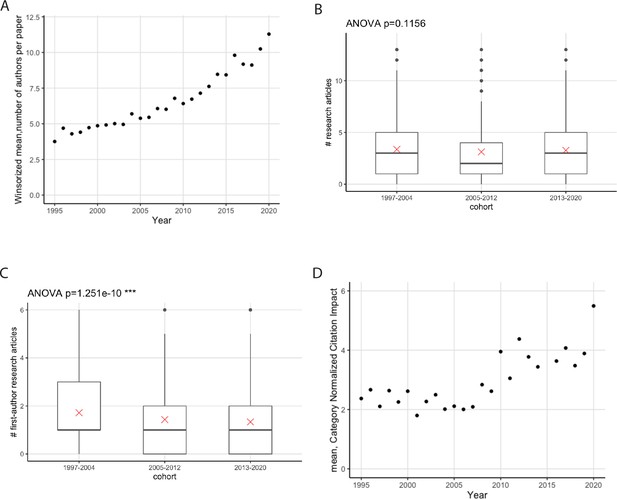
Publications are increasingly collaborative.
( A ) Mean number of authors (y-axis) as a function of year (x-axis) for research articles that were published between 1995 and 2020, and have at least one of the alumni included in this study as an author (n=5413); the winsorized mean has been used to limit the effect of outliers. The mean number of authors has increased by a factor of more than two between 1995 and 2000. ( B ) Boxplot showing the distribution of the number of articles published per researcher for three cohorts. The mean is indicated as a red cross; the circles are outliers. No statistically significant difference was found between the cohorts; the p-value of 0.1156 was generated using a one-way analysis of variance (ANOVA) test of the full dataset (including outliers); the p-value excluding outliers is 0.26. ( C ) Boxplot showing the distribution of the number of first-author articles published per researcher for three cohorts; the two most recent cohorts published fewer first-author articles than the 1997–2004 cohort; the p-value (excluding outliers) was 6.5x10 –7 ; see Table S12 in Supplementary file 1 . ( D ) Mean CNCI (y-axis) as a function of time (x-axis) for research articles that were published between 1995 and 2020, and have at least one of the alumni included in this study as an author (n=5413). Recent articles have higher CNCI values. For clearer visualization, and to protect the identity of alumni with outlying numbers of publications, the y-axis in ( B ) and ( C ) is truncated at the 97.5 percentile.
Figure 5—source data 1
Summary data plotted in Figure 5 .
Many early-career researchers are employed on fixed-term contracts funded by project-based grants, sometimes for a decade or more ( OECD, 2021 ; Acton et al., 2019 ), and surveys suggest that early-career researchers are concerned about career progression ( Woolston, 2020 ; Woolston, 2019 ). We hope PhD students and postdocs will be reassured to learn that the skills and knowledge they acquire during their training are useful in a range of careers both inside and outside acaemica.
Further changes to the career landscape in the life sciences are likely in future, not least as a result of the long-term impacts of the COVID-19 pandemic ( Bodin, 2020 ). It is essential, therefore, that early-career researchers are provided with opportunities to reflect on their strengths, to understand the wide range of career options available to them, and to develop new skills.
The provision of effective support for PhD students and postdocs will require input from different stakeholders – including funders, employers, supervisors and policy makers – and the engagement of the early-career researchers themselves. At EMBL, a career service was launched in 2019 for all PhD students and postdocs, building on a successful EC-funded pilot project that offered career support to 76 postdocs in the EMBL Interdisciplinary Postdoc Programme. The EMBL Fellows’ Career Service now offers career webinars and a blog to the whole scientific community as well as additional tailored support for EMBL PhDs and postdocs including individual career guidance, workshops, resources and events. Funders and policymakers may also need to reassess the sustainably of academic career paths, and to review how funding is allocated between project-based grants and mechanisms that can support PI and non-PI positions with longer-term stability. These measures will will also support equality, diversity and inclusion in science, particularly if paired with research assessment practices that consider factors that can affect apparent research productivity such as career breaks, teaching and service activities.
Factors related to publication are highly predictive of entry into PI careers, and one challenge for an early-career researcher hoping to pursue such a career is to balance the number of articles they publish with the subjective quality of these articles. The trend towards fewer first-author articles per researcher likely reflects a global trend towards articles with more authors and a greater focus on collaborative and/or interdisciplinary approaches to research. Working on a project that involves multiple partners provides an early-career researcher with the opportunity to develop a range of skills, including teamwork, leadership and creativity. Such projects also allow researchers to tackle challenging biological questions from new angles to advance in their field of research, something viewed very positively by academic hiring committees ( Hsu et al., 2021 ; Clement et al., 2020 ; Fernandes et al., 2020 ); however, multi-partner interdisciplinary projects can also take longer to complete. It is therefore important that early-career researchers and their supervisors discuss the potential impact and challenges of (prospective) projects, and what can be done to reduce any risks. For example, open science practices – including author credit statements, FAIR data, and pre-printing – can make project contributions more transparent and available faster ( Kaiser, 2017 ; McNutt et al., 2018 ; Wilkinson et al., 2016 ; Wolf et al., 2021 ).
Limitations
The limitations of our study include that its retrospective, observational design limits our ability to disentangle causation from correlation. The changes in career outcomes may be driven primarily by increased competition for PI roles, but they could also be influenced by a greater availability or awareness of other career options. EMBL has held an annual career day highlighting career options outside academia since 2006, and many of our alumni decide to pursue a career in the private sector, attracted by perceptions of higher pay, more stable contracts, and/or better work-life balance. Likewise, early-career researchers with an interest in a specific technology might, for example, prefer to work at a core facility.
Additionally, we cannot exclude the possibility that other factors may also affect the differences we see between cohorts (such as variations in the number of alumni taking up academic positions in countries that offer later scientific independence). Finally, although comparisons with data from the US and Canada suggest that the trend towards fewer alumni becoming PIs is a global phenomenon, it is possible that some of the trends we observe are specific to EMBL.
We plan to update our observational data every four years, and to maintain data on the career paths of alumni for 24 years after they leave EMBL. This will help us to identify any further changes in the career landscape and to better understand long-term career outcomes in the life sciences. Silva et al., 2019 have also described a method for tracking career outcomes on a yearly basis with estimations of the time and other resources required. We encourage institutions to consider whether they can adapt our methods, or Silva’s method, to the administrative processes and data-privacy regulations applicable at their institutions.
Future studies should also ideally include mixed-method longitudinal studies, which would allow information on career motivations, skills development, research environment, job application activity and other factors to be recorded. Combining the results of such studies with data on career outcomes would allow multifactorial and complex issues, such as gender differences in career outcomes, to be investigated, and would also provide policymakers with a fuller picture of workforce trends. Such studies would, however, require multiple institutions to commit to supplying large amounts of data every year, and coordinating the collection and analysis of such data year-on-year would be a major undertaking that would require the support of funders and institutions.
Data collection and analysis
The study includes individuals who graduated from the EMBL International PhD Programme between 1997 and 2020 (n=969), or who left the EMBL postdoc programme between 1997 and 2020 after spending at least one year as an EMBL postdoctoral fellow (n=1315). Each person is included only once in the study: where a PhD student remained at EMBL for a bridging or longer postdoc, they were included as PhD alumni only, with the postdoc position listed as a career outcome.
For each alumnus or alumna, we retrieved demographic information from our internal records and identified publicly available information about each person’s career path (see Supplementary file 2 ). Where possible, this information was used to reconstruct a detailed career path. An individual was classified as having a "detailed career path" if an online CV or biosketch was found that accounted for their time since EMBL excluding a maximum of two one-calendar-year career breaks (which may, for example, reflect undisclosed sabbaticals or parental leave). Each position was classified using a detailed taxonomy, based on a published schema ( Stayart et al., 2020 ), and given a broad overall classification (see Supplementary file 2 ). The country of the position was also recorded. For the most recent position, we noted whether the job title was indicative of a senior or management level role (i.e., if it included "VP"; "chief"; "cso";"cto"; "ceo"; "head"; "principal”; "president"; "manager"; "leader"; "senior"), or if they appeared to be running a scientific service or core facility in academia.
We use calendar years for all outcome data – for example, for an individual who left EMBL in 2012, the position one calendar year after EMBL would be the position held in 2013. If multiple positions were held in that year, we take the most recent position. We use calendar years, as the available online information often only provides the start and end year of a position (rather than exact date).
An EMBL publication record was also reconstituted for each person in the study. Each of their publications linked to EMBL in the Web of Science and InCites databases in June 2021 were recorded. The data included publication year and – for those indexed in InCites – crude metrics, such as CNCI, percentile in subject area, and journal impact factor. EMBL publications were assigned to individuals in the study based on matching name and publication year (see Supplementary file 2 for full description). When an individual was the second author on a publication, we manually checked for declarations of co-first authorship. Aggregate publication statistics for individuals with the same primary supervisor were also calculated.
The names and other demographic information that would allow easy identification of individuals in the case of a data breach were pseudonymised. A file with key data for analysis and visualisation in R was then generated. A description of this data table can be found in Table S1 in Supplementary file 1 , along with summary statistics.
A Cox proportional hazards regression model was fitted to the data in order to predict time-to-event probabilities for each type of career outcome based on different covariates including cohort, publication variables and gender. Multivariate Cox models were fitted using a ridge penalty with penalty parameter chosen by 10-fold cross-validation. Harrell’s C-index was calculated for each fit in an outer cross-validation scheme for validation and analysis of different models, with 10-fold cross-validation.
The data were collated for the provision of statistics, and are stored in a manner compliant with EMBL's internal policy on data protection . This policy means that the full dataset cannot be made publicly available (because the nature of the data means that sufficient anonymisation is not possible). Summary statistics for the main data table can be found in Supplementary file 1 (Table S1). Rmarkdown documentation of the analysis and figures can be found here and is available on GitHub (copy archived at Coulthard and Lu, 2022 ).
- Toseland CP
- Twelvetrees A
- Google Scholar
- Gilliland G
- Hrabowski FA
- Schlissel MS
- Coulthard R
- Council for Doctoral Education
- Cyranoski D
- Gastelum JB
- Vanderford NL
- Larivière V
- Fernandes JD
- Sarabipour S
- Holehouse AS
- Bisson Filho AW
- Fuhrmann CN
- O’Sullivan PS
- Lindstaedt B
- Rezai-Zadeh KP
- Tennekoon MS
- Markianidou P
- Zacharewicz T
- Threlfall CG
- Cipriano MF
- Golightly LM
- van den Besselaar P
- Slipersæter S
- van der Meulen B
- De Beuckelaer A
- Van der Heyden J
- Martinez ED
- Stavreva DA
- Cohen-Fix O
- McKiernan EC
- Schimanski LA
- Muñoz Nieves C
- Jamieson KH
- Swaminathan S
- Reithmeier R
- Abdulkarim A
- Brouillard L
- Sauermann H
- Schillebeeckx M
- Stanford Biosciences
- Stanford IT&DS
- Flores-Kim EN
- Monsalve GC
- Tregellas JR
- UCSF Graduate Division
- University of Chicago
- University of Michigan
- University Toronto
- Vermeulen N
- von Bartheld CS
- Houmanfar R
- Wilkinson MD
- Dumontier M
- Aalbersberg IJJ
- da Silva Santos LB
- Gonzalez-Beltran A
- ’t Hoen PAC
- Rocca-Serra P
- van Schaik R
- van der Lei J
- van Mulligen E
- Waagmeester A
- Wittenburg P
- Wolstencroft K
- Cossette M-L
- Zerhouni EA
- Peter Rodgers Senior and Reviewing Editor; eLife, United Kingdom
- Barbara Janssens Reviewer; DKFZ (German Cancer Research Center), Heidelberg, Germany
- Sarvenaz Sarabipour Reviewer; Johns Hopkins University, Baltimore, United States
- Reinhart Reithmeier Reviewer; University of Toronto, Toronto, Canada
In the interests of transparency, eLife publishes the most substantive revision requests and the accompanying author responses.
Decision letter after peer review:
Thank you for submitting your article "Meta-research: The changing career paths of PhDs and postdocs trained at EMBL" to eLife for consideration as a Feature Article. Your article has been reviewed by three peer reviewers, and the evaluation has been overseen by the eLife Features Editor (Peter Rodgers). The three reviewers have agreed to reveal their identity: Barbara Janssens; Sarvenaz Sarabipour; Reinhart Reithmeier.
The reviewers and editors have discussed the reviews and we have drafted this decision letter to help you prepare a revised submission. Please also note that eLife does not permit figures in supplementary materials: instead we allow primary figures to have figure supplements. I can suggest how to accommodate the figures in your supplementary materials if your article is accepted.
The eLife Features Editor will also contact you separately about some editorial issues that you will need to address.
This paper deals with career outcomes for some 2284 PhD graduates and post-doctoral fellows from the European Molecular Biology Laboratories, a prestigious research institute that attracts top talent from around the world – the first of its kind. The paper is rich in data beyond simple career outcomes over time and includes gender, publications and collaboration. The methodology involved internet searches like other studies and was enhanced by robust statistical analyses. This important and timely study fills a gap in our knowledge, highlights the important role that institutions like EMBL play in training the next generation of researchers and innovators, and may stimulate other universities and research institutes to do the same. However, there are a number of points that need to be addressed to make the article suitable for publication.
Essential revisions:
1. It is a great achievement to show career destinations for 89% of the 2284 searched for. However the question is in which cohort the 249 "unknown" belong: it could be more transparent to keep those numbers included also in the detailed cohort analysis. It is also unclear, whether the cohort sizes reflect the actual number of researchers who left EMBL in that time period or whether data were lacking. The cohorts reported here increased about 33% from 2004 to 2012 and another 9% by 2020 (Table 2). Could it be, that for more recent cohorts more data are available – for example due to the fact that younger researchers can be found on social media like LinkedIn?
2. The main and interesting conclusion in the abstract is that of the 45% of alumni not continuing in academic research, one third does industry research and one third is in a science-related profession. Other interesting take-home messages are, that a large proportion of alumni changing sectors enter – or are quickly promoted to – managerial positions, that of those who entered a research position in industry one in ten returned to academia and that female Alumni were found less frequently in PI roles. It would be interesting to know, whether there is a difference in these numbers between cohorts – e.g. if more alumni return from industry to academia in recent cohorts and whether female researchers stay longer in postdoc roles (which could influence the total number of female PIs at a given time point).
3. In the time-resolved analysis the authors claim that the probability of being found as a PI in Academia diminished by about 15% after the first cohort (Figure 2C). However the question is whether the absolute number of PIs also decreased. This could be clarified with some reference numbers in the supplementary materials. When calculating the % of the given cohort sizes at different institutes (supp. Table 4) some differences (increase vs decrease) in the number of PIs can be found. Even if the hypothesis that the differences between cohorts are not EMBL-specific, and reflect a wide-spread change in the number of PhDs and postdocs relative to available PI positions seems valid, it would be good to clarify this issue.
4. Interesting findings is also the significant change in time between PhD and first PI position of 0.8 calendar years between the 2004 and 2012 cohorts. It is not surprising that publication factors are highly correlated with entry into PI positions: indeed all ECRs who became PIs published well, but, not all ECRs who published well became PIs! Publications have become more collaborative over the last decade (the number of coauthors has doubled and the number of first author publications per ECR diminished). Another relevant observation is the lack of correlation with group leader seniority or nationality. Group publications were also predictive for other research and science-related careers. Finally a strong observation is that 45% of leavers from the last 5-years who were found to be working outside of academia held senior or management-level roles. These findings can be reassuring for ECRs and the authors could consider to clearly state these in their conclusions.
5. Regarding the career tracking method used for this study: doing google searches for 2284 Alumni is a plausible effort and has probably been time consuming. A question for other research institutes and universities would be, whether this method of career tracking is scalable and/or feasible to continue as a regular task, or whether the authors see this as a one-time effort. If so, what kind or extent of career tracking would the authors recommend to continue sustainably? Performing career tracking is quite relevant, as institutes worldwide now start to be asked to deliver such data to governments and funding agents.
6. In Discussion and future work, it would be valuable to briefly discuss/aspire that institutions such as EMBL compile and publicly report on this type of data/records analysis together with surveys (what authors call mixed methods) of career intent and research environment.
i. With surveys one could have the number of EMBL trainees that actually applied for PI jobs.
ii. The fact that women were found less frequently in PI jobs does not reveal if (1) women apply less frequently or (2) search committees offer PI jobs less often to women or (3) combination of the two.
iii. Surveys together with the data presented in this work can examine the role of lab environment during training and job application.
7. An observation that authors have in the manuscript is that women are less represented as AcPIs (academic PIs). But it's not possible to claim it's an active mechanism. It would be valuable if authors plot the timeline by gender so readers could see the noise.
8. In various panels of Figures 2-4, please clarify if "Time after EMBL" (the label on the x-axis) means "Time after leaving EMBL" or "Time after arrival at EMBL".
Also, it appears that regardless of the cohort, postdocs have increased chances to become a PI only years after they leave EMBL, why? did they go on to do a second postdoc?
9. Please discuss supplementary table 4 in the text, and highlight any common findings from these studies.
10. The authors may also wish to comment on how some members of faculty recruitment committees may need to be trained to recognize bias in relying too heavily on citation indices and first author publications in hiring decisions rather than the scientific contribution of highly-qualified candidates to collaborative projects.
Essential revisions: 1. It is a great achievement to show career destinations for 89% of the 2284 searched for. However the question is in which cohort the 249 "unknown" belong: it could be more transparent to keep those numbers included also in the detailed cohort analysis. It is also unclear, whether the cohort sizes reflect the actual number of researchers who left EMBL in that time period or whether data were lacking. The cohorts reported here increased about 33% from 2004 to 2012 and another 9% by 2020 (Table 2). Could it be, that for more recent cohorts more data are available – for example due to the fact that younger researchers can be found on social media like LinkedIn?
Thank you for these comments, we are happy to clarify here and in the manuscript:
The cohort sizes reflect the number of all PhD students and postdoctoral fellows who left in that time period, according to the official institutional administrative records. The organization grew during the time period included in the study (1997-2020), and the number of PhD students and postdocs in the later cohorts is therefore greater. It excludes individuals who were marked as deceased in our alumni records at the point the data was originally shared with us (June 2017 for 1997-2016 leavers and April 2021 for 2017-2020 leavers; or whose death we learned of during the update of our career tracking data in summer 2021 (through an updated alumni record, or if we found an online obituary)).
For each PhD or postdoc cohort, the percentage of alumni whose current position is unknown is between 9% and 14%, with no consistent trends with time. However, for the oldest cohort we less frequently found a complete online CV or biosketch that was detailed enough to confirm the type of position held for the full-time span since EMBL, particularly for postdoc alumni. Fewer of the older cohort therefore have a detailed CV/career path and there is a higher percentage of unknowns for specific timepoints after EMBL.
Changes to manuscript:
We have added an additional row with the number and percentage of alumni with detailed career paths for each cohort in Table 2 – new row = n(%) detailed career path
We have also clarified that the increased cohort size is due to growth in sentence that refers to this table: “More recent cohorts were also larger (Table 2), reflecting growth of the organization between 1997 and 2020.”
Column charts showing type of position by cohort (Figure 1B and Figure 1 —figure supplement 2 as 1B, but for other time-points) now include all alumni, not just those with a full career path available.
2. The main and interesting conclusion in the abstract is that of the 45% of alumni not continuing in academic research, one third does industry research and one third is in a science-related profession. Other interesting take-home messages are, that a large proportion of alumni changing sectors enter – or are quickly promoted to – managerial positions, that of those who entered a research position in industry one in ten returned to academia and that female Alumni were found less frequently in PI roles. It would be interesting to know, whether there is a difference in these numbers between cohorts – e.g. if more alumni return from industry to academia in recent cohorts and whether female researchers stay longer in postdoc roles (which could influence the total number of female PIs at a given time point). ‘ if more alumni return from industry to academia in recent cohorts’
This is difficult to assess due to the different career lengths and small numbers transitioning from one type of career to another. For example, of 415 alumni who we have a career path for and had at least one Industry role, just 22 returned to a faculty position. Twelve of these were from the oldest cohort (of 117 who held an industry role), compared to 7 (of 178) for the most recent cohort. Similarly, for PI to industry, 21 transitions were observed from the 539 career paths – 16 from the oldest cohort (from 231) and 1 (from 128) in the newest. Given that the propensity to transition may also change with career length, it is difficult to make comparisons or detect meaningful trends from these small numbers.
“whether female researchers stay longer in postdoc roles (which could influence the total number of female PIs at a given time point).”
We did not observe a statistically significant difference in length between PhD and becoming a PI, but agree that his is interesting and that it should be included in the manuscript.
To add this to the manuscript we made three changes: Additional figure supplement showing the average PhD to PI length for male vs female alumni [as previous figure comparing 2005-2012 and 1997-2004 cohorts, but now comparing female vs male alumni] (Figure 3 —figure supplement 1) – this suggests that male and female researchers spend similar times in postdoc roles as the differences are not statistically significant.
We have now included Kaplan Maier plots by gender, which also illustrate the entry into PI (and other) roles with time (as Figure 3B -C and Figure 3 —figure supplement 2 additional plots for AcOt, IndR, NonSci).
We also expand discussion of these data in the main text – see below with new detail italicised. (note: to allow more detailed discussion without requiring repetition of the information, we have moved this section after the sections on changes in career outcomes, where the Kaplan-Meier and PhD to PI length are first discussed).
Many studies have reported that female ECRs are less likely to remain in an academic career (44, 45). Consistent with these previous studies, we observed that male alumni were found more frequently in PI roles (Figure 3A-B; Table S5 in Supplementary information). Figure 3A-B; Table S5 in Supplementary information. There was no statistically significant difference in the time to obtain a PI role between male and female alumni for alumni from 1997-2012, who became PIs within 9-years (Figure 3—figure supplement 1). The difference in career outcomes is therefore unlikely to be explained by different career dynamics for male and female alumni.
Female alumni were more frequently found in science-related non-research roles than male alumni (Figure 3A). In our Cox models, there was also a statistically significant difference between genders in entry into science-related non-research roles for postdoc alumni [p = 0.016] (Figure 3C; Table S5 in Supplementary information).
We more frequently found male alumni in industry research and non-science-related roles than their female colleagues (Figure 3A; Figure 3—figure supplement 2B-C). However, a higher percentage of female than male alumni could not be located. As academics are usually listed on institutional websites, often with a historical publication list that allows unambiguous identification, we expect that most alumni who were not located are employed in the non-academic sector. This means that, considering the higher percentage of female alumni with unknown career paths (where non-academic careers are likely over-represented), the true percentage of female alumni in industry research and non-science-related roles is likely higher, and possibly comparable with the percentage of male alumni in these roles.
We have added information on the absolute number of PIs for each group in the supplementary table that collates the data published from other institutions and comparable EMBL data (in original manuscript table 4; now Table S3 in Supplementary file 1). We have also expanded discussion of this table the text in response to comment 9 (see comment 9 below) and include the absolute numbers.
Additional clarification
In the datasets, the number of PhDs trained per year has increased with time at all institutions. The cohort size – and how much this has increased for more recent cohorts however varies – for example, Stanford’s 2011-2015 cohort was just 18% larger than its 2000-2005 cohort (503 vs 426), whilst the University of Toronto’s 2012-2015 is 96% larger than its 2000-2003 (1234 vs 629). For EMBL, the PhD cohort sizes increased 45% from 256 for the 1997-2004 cohort, to 372 for 2013-2020. Therefore, the absolute number of PIs with time is difficult to compare between institutions. We therefore feel that the percentage of alumni entering different career options is the most pragmatic measure for comparing how career outcomes are changing with time. It can be viewed as the ‘chance’ of a ECR from a specific programme of entering that career area. If an institution continues to train the same absolute number of PIs per year, but trained many more scientists, it nevertheless saw a big difference the career outcomes of its alumni, with more alumni entering non-PI roles.
We have made the following changes to the manuscript to emphasise the positive aspects of our career findings (but balance the editorial comment that “If possible please shorten the first paragraph of the discussion and avoid any unnecessary repetition of material from earlier in the article.”).
This now reads:
“Many ECRs are employed on fixed-term contracts funded by project-based grants, sometimes for a decade or more (52, 53), and surveys suggest that ECRs are concerned about career progression (18-22). We hope ECRs will be reassured by the results of our time-resolved analysis that indicate that the skills and knowledge developed as an ECR can be applied in academia, industry and other sectors. Within academic research, service and teaching, we observed a marked reduction in the percentage of alumni entering PI roles; nevertheless, academic careers continue to be an important career destination. The percentage of alumni entering career areas outside academic research, service and teaching has increased, and our data suggest that ECRs’ skills are valued in these careers; 45% of leavers from the last 5-years who were found to be working outside of academic research and teaching held senior or management-level roles.”
We have expanded the Discussion section ‘Future career studies’ to include a recommendation as follows:
“Evaluating the outcomes of training programmes, and making these data transparently available is a valuable exercise that can provide information to policymakers, transparency for ECRs, and help institutions provide effective career development support. We plan to update our observational data every four years, and maintain data on the career paths of alumni for 24 years after they leave EMBL. This will help us to identify any further changes in the career landscape and better understand long-term career outcomes. Silva et al. (2019) (73) have also described a method for completing career outcome tracking on a yearly basis with estimations of the time and other resources required. We encourage institutions to consider whether they can adapt our, or Silva’s method to the administrative processes and data-privacy regulations applicable at their institutions.”
6. In Discussion and future work, it would be valuable to briefly discuss/aspire that institutions such as EMBL compile and publicly report on this type of data/records analysis together with surveys (what authors call mixed methods) of career intent and research environment. i. With surveys one could have the number of EMBL trainees that actually applied for PI jobs. ii. The fact that women were found less frequently in PI jobs does not reveal if (1) women apply less frequently or (2) search committees offer PI jobs less often to women or (3) combination of the two. iii. Surveys together with the data presented in this work can examine the role of lab environment during training and job application.
Changes to manuscript: We have re-written the ‘Future career studies’ section of the discussion to incorporate these points. This now reads:
“Future studies should also ideally include mixed-method longitudinal studies. This would allow career motivations, skills development, research environment, job application activity, and other factors to be recorded by surveys during ECR training. Correlating these factors to career and training outcomes would allow investigation of multifactorial and complex issues such as gender differences in career outcomes, and provide policymakers with a fuller picture of workforce trends. Such studies will, however, require large sample sizes from multiple institutions and would need significant time investment and coordination over a long time period. The commitment and the support of funders and institutions would therefore be critical.”
Changes to manuscript: We have generated Kaplan Maier plots by gender for entry into each career area, and include these in Figure 3 and Figure 3 —figure supplement 2 (see discussion of reviewer comment 2, above). The hazard ratios, 95% confidence intervals and p values are provided in a supplementary table in file 1 so that the confidence can be judged.
We have added a clarification in the figure legend (“Time after EMBL refers to the number of calendar years between defence of an EMBL PhD and first PI role (for PhD alumni)) – or number of calendar years between leaving the EMBL postdoc programme and first PI role (for postdoc alumni)”, or clarified this in the figure labels, for each figure.
We have expanded on this in the Results section ‘EMBL alumni contribute to research and innovation in academic and non-academic roles’, adding the following text:
“On average, PhD alumni became PIs 6.1 calendar years after their PhD defence. For postdoc alumni, the start year of the first PI role was on average 7.3 calendar years after their PhD and 2.5 years after completing their EMBL postdoc. Almost half of EMBL postdoc alumni who became PIs did so directly after completing their EMBL postdoc (168 of 343 alumni with a detailed career path available). Other postdoc alumni made the transition later, most frequently after one additional postdoc (71 alumni) or a single academic research / teaching / service position (56 alumni). Some alumni held multiple academic (40 alumni), or one or more non-academic positions (8 alumni) between their EMBL postdoc and first PI role.”
And to provide balance / to avoid focusing only on academic careers, we also include an additional sentence in the subsequent paragraph on non-academic areas:
“The average time between being awarded a PhD and the first industry research, science-related or non-science-related role was 5.0, 5.3 and 4.2 calendar years respectively.”
We have expanded the discussion of this in the Results section ‘The percentage of EMBL alumni becoming PIs is similar to data released by North American institutions for both older and more recent cohorts’. [note that due to rearrangements, supplementary table 4 is now ‘Table S3 in Supplementary file 1’]. This now reads:
“A number of institutions have released cohort-based PhD outcomes data online or in publications (32, 44-49). Of these, a recent dataset from Stanford University offers the longest career tracking, reporting outcomes for PhD graduates from 20 graduation years (2000-2019) (44). In this dataset, Stanford University reported that 34% (145/426) of its 2000-2005 PhD alumni were in research-focussed faculty roles in 2018. This is comparable to the 37% (78/210) we observe for the EMBL alumni for the same time period. For 2011-2015 graduates, comparable percentages of Stanford (13%, 63/503) and EMBL (11%, 25/234) graduates were also in PI roles in 2018 (Figure 1C). Figure 1 – Supplement 3 plots data from five other datasets alongside equivalent data from EMBL (further details of each dataset are available in the Table S3 in Supplementary file 1). This includes a published dataset from the University of Toronto (32, 49), which reported that 31% (192/629) of its 2000-2003 life science division graduates and 25% (203/816) of its 2004-2007 life science graduates were in tenure stream roles in 2016. The equivalent EMBL data is comparable at 39% (52/132) and 28% (49/172) of graduates in PI roles respectively. Across all six datasets, EMBL and the other institutes generally have a similar proportion of alumni entering PI roles for comparable cohorts. This is consistent with our hypothesis that the differences between cohorts are not EMBL-specific, and reflect a wide-spread change in the number of PhDs and postdocs relative to available PI positions.”
We have expanded this section of the discussion (third paragraph of ‘Addressing ECR career challenges’ in the discussion), also mentioning the trend to narrative CVs that has accelerated recently. This now reads:
“Research assessment and availability of funding play an important role in determining the career prospects of an academic. Therefore, it is also vital that factors that may lead to misperception of the productivity of ECRs, such as involvement in large-scale projects, career breaks, or time spent on teaching and service, are considered in research assessment during hiring, promotion and funding decisions. Initiatives such as the San Francisco Declaration on Research Assessment (DORA) and Coalition for Advancing Research Assessment (CoARA) have been advocating for an increased focus on good practice in evaluating research outputs, and many institutions and funders have reviewed their practices. Cancer Research UK, for example, now asks applicants to its grants to describe three to five research achievements, which can include non-publication outputs (64) and narrative CV formats that allow candidates to put their achievements in context are also becoming more common (61). The impact of the coronavirus pandemic on research productivity of researchers with caregiving responsibilities makes such actions imperative (65-67). We welcome this increased focus on qualitative assessment of scientific contribution, rather than reliance on publication metrics. To ensure successful implementation and to avoid unintended consequences (such as introducing new biases), it will be important for funders and institutions to provide appropriate guidance and/or training to evaluators and to carefully monitor implementation. Other initiatives that may help ECR involved in ambitious projects to demonstrate their contributions include more transparent author contribution information in publications (68, 69) and promotion of "FAIR" principles of data management (70). The increasing use of preprints (41, 71) is also likely to have a positive effect on the careers of ECRs involved in projects with longer time scales (72).”
Author details
Junyan Lu , Genome Biology Unit, European Molecular Biology Laboratory, Heidelberg, Germany
Present address
Contribution, competing interests.
Britta Velten , Genome Biology Unit, European Molecular Biology Laboratory, Heidelberg, Germany
Contributed equally with
Bernd Klaus , Genome Biology Unit, European Molecular Biology Laboratory, Heidelberg, Germany
Mauricio Ramm , EMBL International Centre for Advanced Training, European Molecular Biology Laboratory, Heidelberg, Germany
Wolfgang Huber , Genome Biology Unit, European Molecular Biology Laboratory, Heidelberg, Germany
Rachel Coulthard-Graf , EMBL International Centre for Advanced Training, European Molecular Biology Laboratory, Heidelberg, Germany

For correspondence
Horizon 2020 framework programme (664726), horizon 2020 framework programme (847543), european molecular biology laboratory.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
We thank Monika Lachner and Anne Ephrussi for their critical reading of the manuscript and strong support of this project. We also acknowledge the instrumental support of the Alumni Relations, DPO, HR, SAP, Library, International PhD Programme and Postdoc Programme teams at EMBL. We also thank Edith Heard, Brenda Stride, Jana Watson-Kapps (FMI), and the Directorate, SAC, SSMAC and Council of the EMBL for discussion. The work was supported by: EMBL (JL, BK, MR, WH, RCG) and the EMBL International PhD Programme (BV). RCG is employed by EMBL’s Interdisciplinary Postdoc Programme, which has received funding from the European Union’s Horizon 2020 programme (Marie Skłodowska-Curie Actions).
Publication history
- Preprint posted: March 1, 2022 (view preprint)
- Received: March 17, 2022
- Accepted: November 8, 2023
- Version of Record published: November 23, 2023 (version 1)
© 2023, Lu et al.
This article is distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use and redistribution provided that the original author and source are credited.
- 8,427 Page views
- 255 Downloads
- 1 Citations
Article citation count generated by polling the highest count across the following sources: Crossref , PubMed Central , Scopus .
Download links
Downloads (link to download the article as pdf).
- Article PDF
- Figures PDF
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools), categories and tags.
- workforce trends
- graduate school
- early-career researcher
- careers in science
- Part of Collection

Meta-Research: A Collection of Articles
Further reading.
The study of science itself is a growing field of research.
Listen to Rachel Coulthard-Graf talk about career paths for scientists.
An analysis of the career paths of more than 2000 PhDs and postdocs provides insights into the evolving career landscape for life scientists
Be the first to read new articles from eLife

- Postdoc India
- Postdoc Abroad
- Postdoc (SS)
- RESEARCHERSJOB
- Post a position
- JRF/SRF/Project
- Science News

PhD Positions in Life Sciences, European Molecular Biology Laboratory (EMBL)

PhD Positions in Life Sciences: The European Molecular Biology Laboratory (EMBL) is offering exciting PhD Positions across its sites in Germany, UK, France, Italy, Spain. Join an international team of scientists contributing to basic research in molecular life sciences.
Summary Table:
Study Area: PhD Positions in various locations (Heidelberg, Barcelona, Grenoble, Hamburg, Hinxton, Rome) focusing on molecular life sciences. Explore research topics at EMBL here .
Scholarship Description: EMBL provides a starting platform for a successful scientific career through early independence and interdisciplinary research. The program offers fully funded PhD positions with comprehensive health care and pension benefits.
Eligibility: Applicants with diverse backgrounds in Biology, Chemistry, Physics, Mathematics, Computer Science, Engineering, and Molecular Medicine are welcome. Candidates must hold or anticipate receiving a qualifying university degree before enrollment in October of the given year.
Required Documents:
- Online application link
- References (submitted by 13 March 2024)
How to Apply: Learn more about the EMBL International PhD Programme and apply online here . The application deadline is 11 March 2024 . Interviews are scheduled for June 2024, and successful candidates will start their work at EMBL by mid-October 2024.
Last Date: Don’t miss the opportunity to be part of groundbreaking research in molecular life sciences. The deadline for submitting online applications is 11 March 2024 . For further information, contact EMBL Graduate Office via [email protected] . Join EMBL and make your PhD journey a formative and enriching experience!

RELATED ARTICLES MORE FROM AUTHOR
Phd position in hydrogen storage, heriot-watt university, united kingdom, fully-funded phd studentship in environmental planning at swansea university, uk, phd position in ai-based analysis, tu/e, eindhoven, netherlands, doctoral researcher in computational biophysics, university of helsinki, finland, three phd positions, utrecht university, netherlands, two phd students: energy systems analysis, chalmers university of technology, sweden, leave a reply cancel reply.
Save my name, email, and website in this browser for the next time I comment.
Follow us on Instagram @researchersjob_rj
- Terms Of Service
- Privacy Policy
Postdoc: Antigen Interactions, Netherlands Cancer Institute (NKI)

EMBL Events
EMBL delivers world-class courses, conferences and workshops at the forefront of molecular life science and its applications.
Browse upcoming events below, or learn more about training at EMBL.
8 April - 12 April 2024
scATAC-seq: attacking open chromatin in single cells
Vladimir Benes, Ulrike Litzenburger, Judith Zaugg
ATAC Seq stands for Assay of Transposase Accessible Chromatin and is a widespread method to profile open chromatin on a genome scale The goal of this introductory course is to teach the entire workflow of a single cell and bulk ATAC Seq experiment including library preparation sequencing data processing and downstream analysis by addressing a real biological question that will be answered at the end of the course... OverviewSpeakersProgrammePractical informationSponsorsMedia kit Overview Course overview ATAC-Seq stands for Assay of Transposase Accessible Chromatin and is a widespread method to profile open chromatin on a genome-scale. The goal of this introductory course is to teach the entire workflow of a single cell ATAC-Seq experiment including library preparation, sequencing, data processing, and downstream analysis by addressing a real biological question that will be answered at the end of the course. We will start with the experimental single cell ATAC-Seq protocol, highlighting the critical steps, potential pitfalls, and the most up-to-date tweaks to improve the quality of the results. After sequencing the samples we will go through the basic quality control and preprocessing steps using multiple analysis tools for next generation sequencing data. Finally, we will perform different types of...
EMBL Heidelberg
Data-driven approaches to understanding dementia
Juanita Riveros
This course provides individuals working in dementia research with the skills to access and analyse dementia data Participants will learn to apply bioinformatics methodologies combine different data types and utilise relevant dementia data resources enhancing their research capabilities in the field... This course provides individuals working in dementia research with the skills to access and analyse dementia data. Participants will learn to apply bioinformatics methodologies, combine different data types, and utilise relevant dementia data resources, enhancing their research capabilities in the field.
9 April - 12 April 2024
Diversity of plants: from genomes to metabolism
Barbara Ann Halkier, Sarah E. O’Connor, Detlef Weigel
OverviewSpeakersProgrammePractical informationSponsorsAboutMedia kit Overview This conference will take place at EMBL Heidelberg with the option to attend virtually Symposium overview This inaugural EMBO EMBL Symposium will showcase the future of plant biology Going beyond the limited number of wild and crop plants that have historically been focused on modern comparative approaches... OverviewSpeakersProgrammePractical informationSponsorsAboutMedia kit Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Symposium overview This inaugural EMBO | EMBL Symposium will showcase the future of plant biology. Going beyond the limited number of wild and crop plants that have historically been focused on, modern comparative approaches are revealing the mechanistic bases and evolutionary forces driving the enormous phenotypic variation among the 400,000 plant species on Earth. This conference is for biologists with an interest in plant diversity at the genomic, (epi)genetic, phenotypic and metabolic level. The major topical threads will be the biology of genomes, evolution of metabolic pathways, and evolution of environment-dependent and -independent phenotypes, which constitute some of the most exciting areas of contemporary biology....
EMBO | EMBL Symposium
EMBL Heidelberg, Virtual
11 April 2024
Curating proteins involved in Antimicrobial Resistance (AMR) in UniProt
UniProt is a comprehensive expert led publicly available database of protein sequence function and variation information This webinar will give a brief introduction to the UniProt website and then highlight resources which should be of interest to people beginning work on antimicrobial resistance AMR including students early stage researchers and clinicians AMR constitutes one of the world s most urgent public health problems We have been improving the annotation of various protein classes with a known role in AMR including beta lactamases efflux pumps ABC transporters and a group of proteins essential for the survival of resistant bacteria under therapeutic concentrations of antimicrobials collectively known as the secondary resistome Using these as examples we will introduce some of the UniProt resources UniProt annotations are widely disseminated to other databases including Rhea EnsemblBacteria PATRIC Bacterial Viral Bioinformatics Resource Center BioCyc EcoCyc and KEGG Please also take this opportunity to suggest additional curation targets to us to enable us to better support ongoing AMR research... UniProt is a comprehensive, expert-led, publicly available database of protein sequence, function, and variation information.This webinar will give a brief introduction to the UniProt website and then highlight resources which should be of interest to people beginning work on antimicrobial resistance (AMR), including students, early-stage researchers, and clinicians.AMR constitutes one of the world’s most urgent public health problems.We have been improving the annotation of various protein classes with a known role in AMR, including beta-lactamases, efflux pumps, ABC transporters, and a group of proteins, essential for the survival of resistant bacteria under therapeutic concentrations of antimicrobials, collectively known as the secondary resistome. Using these as examples, we will introduce some of the UniProt resources.UniProt annotations are widely disseminated to other databases,...
15 April - 19 April 2024
Ultrastructure expansion microscopy
Niccolo Banterle, Gautam Dey, Paul Guichard, Virginie Hamel, Marine Laporte
Expansion microscopy ExM physically magnifies specimens allowing to obtain super resolution images using a conventional diffraction limited microscope such as confocal microscopy This powerful technique is easy to implement but it requires some hands on training... OverviewSpeakersProgrammePractical informationSponsorsMedia kit Overview Course overview Expansion microscopy (ExM) physically magnifies specimens, allowing to obtain super-resolution images using a conventional diffraction-limited microscope such as confocal microscopy. This powerful technique is easy to implement but it requires some hands-on training. The gain in resolution can be further enhanced with the use of super-resolution microscopes such as STED. This course centered on the Ultrastructure Expansion Microscopy (U-ExM) based techniques applied to mammalian cells aims at disseminating and discussing expansion microscopy protocols and is intended for cell biologists, parasitologists, etc, with applications ranging from tissue to cells. Audience This course is intended for researchers ranging from PhD students to post-docs and senior scientists as well as staff from bioimaging...
EMBO Practical Course
15 April - 18 April 2024
The mechanics of life: from development to disease
Prisca Liberali, Nicoletta Petridou, Amy Shyer, Andela Saric, Xavier Trepat
OverviewSpeakersPreliminary programmePractical informationSponsorsAboutMedia kit Overview This conference will take place at EMBL Heidelberg with the option to attend virtually Symposium overview The building blocks of life macromolecules cells tissues and organs act on each other via physical forces This interplay of physical forces between the units of life gives rise to complex... OverviewSpeakersPreliminary programmePractical informationSponsorsAboutMedia kit Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Symposium overview The building blocks of life – macromolecules, cells, tissues, and organs – act on each other via physical forces. This interplay of physical forces between the units of life gives rise to complex behaviour — enables motility, transport, growth, division, response to environmental cues, and allows the exertion of forces on the environment, bridging biological scales. The alteration of forces and mechanical properties is associated with inflammation, cancer, and ageing. This comprehensive symposium will study the interplay of the physical forces — the mechanics — between the fundamental building blocks of life, from molecules to organs. It will cover how these physical forces enable development...
16 April - 20 April 2024
Computational Biology Training in Hematology 2024, Module 1
Sophie Spencer
Computational Biology Training in Hematology CBTH provides early career researchers aiming to specialize in computational and quantitative biology aspects of hematology with a unique year long training and mentoring experience by top tier faculty CBTH is a joint effort of the European Hematology Association EHA and the European Molecular Biology Laboratory European Bioinformatics Institute EMBL EBI that aims to support the development of quantitative skills for hematology researchers through training and mentoring in computational biology The CBTH program will be organized with two multi day workshops in a city in Europe and a follow up session at the EHA Congress... Computational Biology Training in Hematology (CBTH) provides early-career researchers aiming to specialize in computational and quantitative biology aspects of hematology with a unique, year-long training and mentoring experience by top-tier faculty.CBTH is a joint effort of the European Hematology Association (EHA) and the European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI) that aims to support the development of quantitative skills for hematology researchers through training and mentoring in computational biology.The CBTH program will be organized with two multi-day workshops in a city in Europe and a follow up session at the EHA Congress.
17 April 2024
Statistical thinking for microbial ecology
Most microbial ecologists understand the importance of rigorous statistical analysis for conducting a replicable study However formal courses in the statistical analysis of high dimensional ecological data are rare and traditional courses in applied statistics may not teach skills applicable to modern microbiome analysis This webinar will introduce foundational concepts in statistics with a focus on applying these concepts to the analysis of modern microbiome datasets To illustrate these concepts we will use the field of differential abundance as a case study We will contrast popular differential abundance parameters and estimators specifically focusing on estimating meaningful contrasts using high throughput sequencing data... Most microbial ecologists understand the importance of rigorous statistical analysis for conducting a replicable study. However, formal courses in the statistical analysis of high-dimensional ecological data are rare, and traditional courses in applied statistics may not teach skills applicable to modern microbiome analysis.This webinar will introduce foundational concepts in statistics, with a focus on applying these concepts to the analysis of modern microbiome datasets. To illustrate these concepts, we will use the field of "differential abundance" as a case study. We will contrast popular differential abundance parameters and estimators, specifically focusing on estimating meaningful contrasts using high-throughput sequencing data.
22 April - 26 April 2024
Microscopy data analysis: machine learning and the BioImage Archive
Meredith Willmott
This virtual course will show how public bioimaging data resources centred around the BioImage Archive enable and enhance machine learning based image analysis The content will explore a variety of data types including electron and light microscopy and miscellaneous or multi modal imaging data at the cell and tissue scale Participants will cover contemporary biological image analysis with an emphasis on machine learning methods as well as how to access and use images from databases Further instruction will be offered using applications such as ZeroCostDL4Mic ilastik the BioImage Model Zoo and CellProfiler Virtual course This course will be a virtual event delivered via a mixture of live streamed sessions pre recorded lectures and tutorials with live support We will be using Zoom to run the live sessions all fully password protected with automated English closed captioning and transcription with support and both scientific and social networking opportunities provided by Slack and other methods taking different time zones into account In order to make the most out of the course you should make sure to have a stable internet connection throughout the week and are available between 08 00 18 00 BST each day In the week before the course there will be a brief induction session Computational practicals will run on EMBL EBI s virtual training infrastructure meaning participants will not require access to a powerful computer or install complex software on their own machines Selected participants may be sent materials prior to the course These might include pre recorded talks and required reading or online training that will be essential to fully engage with the course... This virtual course will show how public bioimaging data resources, centred around the BioImage Archive, enable and enhance machine learning based image analysis. The content will explore a variety of data types including electron and light microscopy and miscellaneous or multi-modal imaging data at the cell and tissue scale. Participants will cover contemporary biological image analysis with an emphasis on machine learning methods, as well as how to access and use images from databases. Further instruction will be offered using applications such as ZeroCostDL4Mic, ilastik, the BioImage Model Zoo, and CellProfiler. Virtual course This course will be a virtual event delivered via a mixture of live-streamed sessions, pre-recorded lectures, and tutorials with live support. We will be using Zoom to run the live sessions (all fully password protected with automated English closed captioning...
Fundamentals of widefield and confocal microscopy and imaging
Heiko Gaethje, Jens Marquard
This course will provide the participants with an overview of the basics of microscopy light and optics contrast techniques and imaging... OverviewSpeakersPreliminary programmePractical informationSponsorsMedia kit Overview Registration is not yet open for this event. If you are interested in receiving more information please register your interest. Course overview Light microscopy is the most widely used form of microscopy for life science research. The correct handling of the microscope and optimising image quality (e.g. contrast, resolution) as well as experimental design according to different applications are crucial to obtain the best results. This course will provide the participants with an overview of the basics of microscopy (light and optics, contrast techniques and imaging). The confocal part will include multicolour, 3D, and time-lapse imaging as well as spectral imaging, introduction to photobleaching and photoactivation techniques. Basic image analysis tools including measurement functions, colocalisation,...
23 April - 26 April 2024
Organismal physiology
Gerard Karsenty, Irene Miguel-Aliaga, Asya Rolls, Miguel Soares
This symposium aims to provide a high visibility platform to scientists from multiple disciplines who use molecular genetic tools to probe inter organ communication signals that maintain homeostasis It will also directly address a fundamental and long term goal of the study of physiology how can recent developments in whole organism physiology pave the way to novel adapted and efficient therapies for multiple degenerative diseases... OverviewSpeakersPreliminary programmePractical informationSponsorsAbout Overview We regret to inform you that this event has been cancelled. Please have a look at our other upcoming events here. Symposium overview “Like a machine, any organism, even the simplest, is a functional unit, coherent and integrated. We are far from having elucidated, in complex organisms, the entire structure of these functional units”. A major outcome of research using model organisms has been to address this central question of biology that was stated so clearly and candidly by J. Monod in 1969. Genetic studies in model organism have first revealed that we are far from knowing all the functions fulfilled by various organs in vertebrates or invertebrates. In doing so, this body of work has nonetheless led to unprecedented progress in our understanding of how homeostasis of an entire organism is maintained. As...
24 April 2024
Introduction to the Ensembl Bacteria genome browser
Ensembl Bacteria is a browser for bacterial and archaeal genomes In this webinar we will explore Ensembl Bacteria to learn more about the latest data and where it came from view annotation of molecular interactions involving bacterial genes and comparative genomics data involving key bacterial species... Ensembl Bacteria is a browser for bacterial and archaeal genomes. In this webinar, we will explore Ensembl Bacteria to learn more about the latest data and where it came from, view annotation of molecular interactions involving bacterial genes and comparative genomics data involving key bacterial species.
Harmony in diversity: exploring the rich microbiomes of South American wildlife
Colombia and northern South America is known as a biodiversity hotspot but at the basis of that diversity is the microbial communities With recent advances in metagenomics the microbial communities and genomic composition has started to be characterised by unveiling a whole new world that is highly interconnected with macro organisms and the environment This webinar will illustrate the intricate world of microbial communities associated with diverse South American fauna From the high altitude realms of neotropical bumblebees to the depths of coral reefs we ll journey through the microbiome landscapes of beings like spectacled bears woolly monkeys lichens shrimps among others Discover the unseen connections shaping the health and resilience of these unique ecosystems... Colombia and northern South America is known as a biodiversity hotspot, but at the basis of that diversity is the microbial communities. With recent advances in metagenomics, the microbial communities and genomic composition has started to be characterised by unveiling a whole new world that is highly interconnected with macro-organisms and the environment.This webinar will illustrate the intricate world of microbial communities associated with diverse South American fauna. From the high-altitude realms of neotropical bumblebees to the depths of coral reefs, we'll journey through the microbiome landscapes of beings like spectacled bears, woolly monkeys, lichens, shrimps, among others. Discover the unseen connections shaping the health and resilience of these unique ecosystems.
6 May - 8 May 2024
Building networks 2024: engineering in vascular biology
Maria Bernabeu, Claudio Franco, Stephan Huveneers, Milica Radisic
In this meeting we intend to bring together vascular biologists and bioengineers from across the world to build research synergies to tackle long standing questions in the field of vascular biology... OverviewSpeakersProgrammePractical informationSponsorsMedia kit Overview This conference will take place at EMBL Barcelona. Conference overview Blood vessels are essential for the formation and maintenance of larger tissues and organs. The vascular system regulates exchange of oxygen, nutrients and immune cells – the dysfunction of which can cause severe pathologies. Mouse models, genetics and histology of human samples, coupled with recent advances in imaging, single-cell approaches and computational modelling, have provided significant knowledge to the field of vascular biology. Yet, in vivo models are highly complex and often need to be combined with simpler in vitro studies to get mechanistic and molecular insights. Growth of in vitro blood vessels presents numerous research challenges due to their complex 3D geometry, and a strong coupling between biology and mechanics.Recent...
EMBO Workshop
EMBL Barcelona
Wine yeasts as a model system in community ecology
Wine fermentation constitutes a transient microbial ecosystem wherein numerous yeast species originating from the grape surface engage in interactions This leads to an ecological succession anticipated to culminate in the ultimate dominance of Saccharomyces cerevisiae The diversity of yeast species found in grape musts has a direct impact on the final chemical and aroma complexity of the resulting wines Understanding the molecular and ecological interaction patterns between wine yeast communities is essential to harness diverse yeast functionalities with the final aim of producing tailored high quality wines In this seminar we will introduce the expectable phylogenetic and functional diversity in wine yeast communities we will discuss how to engineer wine yeast communities top down and bottom up and we will finally conclude about the necessity of adopting the adequate theoretical framework to address challenging questions in applied ecology and to finally improve microbiome based industrial processes... Wine fermentation constitutes a transient microbial ecosystem wherein numerous yeast species, originating from the grape surface, engage in interactions. This leads to an ecological succession anticipated to culminate in the ultimate dominance of Saccharomyces cerevisiae. The diversity of yeast species found in grape musts has a direct impact on the final chemical and aroma complexity of the resulting wines. Understanding the molecular and ecological interaction patterns between wine yeast communities is essential to harness diverse yeast functionalities, with the final aim of producing tailored high-quality wines. In this seminar we will introduce the expectable phylogenetic and functional diversity in wine yeast communities, we will discuss how to engineer wine yeast communities (top-down and bottom-up), and we will finally conclude about the necessity of adopting the adequate...
12 May - 17 May 2024
Advanced fluorescence imaging techniques
Manuel Gunkel, Aliaksandr Halavatyi, Debora Keller-Olivier Joana Delgado Martins, Irina Rakotoson
This practical course will cover advanced light microscopy techniques and participants will learn how to derive qualitative and quantitative insights on molecular mechanisms in cells and developing organisms Invited guest researchers together with experts from the EMBL and microscopy professionals will foster an intense information flow with a balance of lectures and practical workshops The focus of the course will be on the use of fluorescence microscopy to obtain information about protein localisation dynamics and functions at different spatial scales... OverviewSpeakersPreliminary programmePractical informationSponsorsMedia Kit Overview Course overview This practical course will cover advanced light microscopy techniques and participants will learn how to derive qualitative and quantitative insights on molecular mechanisms in cells and developing organisms. Invited guest researchers together with microscopy experts from EMBL will foster an intense information flow with a balance of lectures and practical workshops. The focus of the course will be on the use of fluorescence microscopy to obtain information about localisation and functions of proteins and other bio-molecules at different spatial scales. Audience This course is directed towards researchers in the life sciences with access to light microscopy equipment, who have an immediate need to apply the techniques they will learn to solve biological problems at the cellular and...
EMBL Course
14 May - 17 May 2024
Cellular mechanisms driven by phase separation
Simon Alberti, Sara Cuylen-Häring, Dorothee Dormann, Alex Holehouse
This conference will bring together scientists from soft matter polymer physics molecular biology structural biology and cell and developmental biology to study condensates in biology and disease... OverviewSpeakersPreliminary programmePractical informationSponsorsAboutMedia kit Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Symposium overview Phase separation is emerging as a common biophysical principle underlying many important cellular functions. This conference will bring together scientists from cell biology, biophysics, biochemistry, molecular biology, structural biology, developmental biology, polymer physics, and soft matter physics to study condensates in biology and disease. The goals of this symposium are to: increase our understanding of the physiological relevance of condensates in many biological processes explore the large repertoire of new tools and methods that can be applied to study the phenomenon of biological condensates provide transdisciplinary training to tackle the role of condensates in biology Session...
Introduction to metabolomics analysis
This course will provide an introduction to metabolomics through lectures and hands on sessions using publicly available data software and tools Participants will become familiar with standardised workflows as well as with the current state of experimental design data acquisition LC MS MS imaging processing and modelling In addition they will learn about community standards and sharing in metabolomics particularly through the use of EMBL EBI s MetaboLights repository and Galaxy infrastructure Participants will learn through hands on tutorials to use tools available for data analysis and data submission Additionally case studies will be discussed to show how to employ the week s learning... This course will provide an introduction to metabolomics through lectures and hands-on sessions, using publicly available data, software, and tools. Participants will become familiar with standardised workflows as well as with the current state of experimental design, data acquisition (LC-MS, MS imaging), processing, and modelling. In addition, they will learn about community standards and sharing in metabolomics, particularly through the use of EMBL-EBI’s MetaboLights repository and Galaxy infrastructure. Participants will learn through hands-on tutorials to use tools available for data analysis and data submission. Additionally, case studies will be discussed to show how to employ the week’s learning.
21 May - 23 May 2024
BioMalPar XX: biology and pathology of the malaria parasite
Sabrina Absalon, Jess Bryant, Fredros Okumu, Taco Kooij
OverviewSpeakersProgrammePractical informationSponsorsMedia kit Overview This conference will take place at EMBL Heidelberg with the option to attend virtually Abstract submission for virtual short talks and financial assistance for virtual participation can be submitted by Monday 1 April 2024 Conference overview Malaria remains a major global public health challenge The emergence... OverviewSpeakersProgrammePractical informationSponsorsMedia kit Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Abstract submission for virtual short talks and financial assistance for virtual participation can be submitted by Monday 1 April 2024. Conference overview Malaria remains a major global public health challenge. The emergence and spread of resistance to existing antimalarial drugs further complicates the fight against this ancient foe. As the BioMalPar Conference marks its 20th anniversary, we would like to take a moment to reflect on the critical mission of understanding the biology of the Plasmodium parasite, its interactions with hosts and vectors, and the urgent need to develop effective intervention strategies. This year, we will focus on enhancing our research community by forging new connections. We recognize that...
EMBL Conference
24 May 2024
Interpreting the effects of genetic variants on protein structure and function
Genetic variation between individuals results in a range of human phenotypes Variants that fall within coding regions of genes generally have direct effects on the proteins encoded by the genes with which they overlap This six hour virtual workshop will provide participants with an opportunity to gain hands on experience of using EMBL EBI tools and resources to assess the effects of genetic variants on the structure and function of proteins in a common workflow During the course a single example will be used throughout using different EMBL EBI tools and resources at each stage of the analysis pipeline The example will involve running a VCF file through the Ensembl Variant Effect Predictor VEP then following links to UniProt PDBe and AlphaFoldDB for further variant information and interpretation Virtual courseThis course will take place on Zoom Trainers will be available to assist during practical sessions answer questions and provide further explanations during the Zoom To join the course you will need to create an EMBL EBI Training website account... Genetic variation between individuals results in a range of human phenotypes. Variants that fall within coding regions of genes generally have direct effects on the proteins encoded by the genes with which they overlap. This six-hour virtual workshop will provide participants with an opportunity to gain hands-on experience of using EMBL-EBI tools and resources to assess the effects of genetic variants on the structure and function of proteins in a common workflow. During the course, a single example will be used throughout, using different EMBL-EBI tools and resources at each stage of the analysis pipeline. The example will involve running a VCF file through the Ensembl Variant Effect Predictor (VEP), then following links to UniProt, PDBe, and AlphaFoldDB for further variant information and interpretation. Virtual courseThis course will take place on Zoom. Trainers will be...
26 May - 31 May 2024
Whole transcriptome data analysis
Marco Beccuti, Vladimir Benes, Raffaele Calogero, Francesca Cordero
This course will teach the biological researchers how to analyse biological data sets using open source software Most of the analysis will be performed with docker4seq and rCASC packages which was developed to facilitate the use of computing demanding applications in the field of NGS data analysis... OverviewSpeakersProgrammePractical informationSponsorsMedia kit Overview Course overview This course will teach the biological researchers how to analyse biological data sets using open-source software. Most of the analysis will be performed with docker4seq and rCASC packages, which was developed to facilitate the use of computing demanding applications in the field of NGS data analysis. Docker4seq and rCASC packages use docker containers that embed demanding computing tasks (e.g. short reads mapping) into isolated containers. This approach provides multiple advantages: the user does not need to install all the software on its local server, the results generated by different containers can be organised in pipelines and reproducible research is guarantee by the possibility of sharing the docker images used for the analysis. Computers for hands-on exercises will be provided along with demo...
1 June - 8 June 2024
Structural characterisation of macromolecular complexes
Ana Casanal, Wojciech Galej, Eva Kowalinski, Carlo Petosa, Montserrat Soler López
OverviewSpeakers and trainersPreliminary programmePractical informationSponsorsMedia kit Overview Course overview Recent developments in cryo EM ET super resolution microscopy and protein structure prediction coupled with increasingly sophisticated methods of sample preparation have revolutionised structural biology Educating the next generation of researchers on how to strategically... OverviewSpeakers and trainersPreliminary programmePractical informationSponsorsMedia kit Overview Course overview Recent developments in cryo-EM/ET, super-resolution microscopy, and protein structure prediction — coupled with increasingly sophisticated methods of sample preparation — have revolutionised structural biology. Educating the next generation of researchers on how to strategically combine these disparate tools is critical for addressing the most challenging structural biology problems of the future. Course participants will receive a comprehensive overview of state-of-the-art structural methods, with a focus on sample preparation, characterisation, and data integration strategies. Key aspects covered by lectures and practicals include multi-subunit protein expression in bacteria, insect, and mammalian cells; isolation of native complexes and in-vitro reconstitution of protein...
EMBL Grenoble
5 June - 8 June 2024
Microtubules: from atoms to complex systems
Carsten Janke, Thomas Müller-Reichert, Shinsuke Niwa, Kassandra Ori-Mckenney
In recent years interdisciplinary approaches embracing cell biology genetics molecular biology biochemistry biophysics structural biology and mathematical modelling have made a tremendous impact on the microtubule field... OverviewSpeakersPreliminary ProgrammeAboutPractical infomationSponsorsMedia kit Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Symposium overview This symposium, initiated in 2010, has taken place every second year since and brings together early-career and established researchers from all over the world who are interested in microtubule biology and its impact on human health and disease. The microtubule cytoskeleton is essential for a wide variety of cellular functions, such as chromosome segregation, directed vesicle and organelle transport, cell motility, and cell polarity. Impaired microtubule function can lead to human diseases including cancer and neurodegenerative disorders, where microtubules serve as important therapeutic targets. In recent years, interdisciplinary approaches embracing cell biology, genetics, molecular biology,...
9 June - 15 June 2024
C. elegans: from genome editing to imaging
Henrik Bringmann, Simone Koehler, Georgia Rapti, Pavak Shah
OverviewSpeakersPreliminary ProgrammePractical informationSponsorsMedia kit Overview Course overview C elegans is a popular experimental model that combines transparency reproducible anatomy traceable cell lineage sequenced genome and facile genetics allowing to dissect animal development physiology and behaviour across scales of genes cells and tissues This 7 day course will... OverviewSpeakersPreliminary ProgrammePractical informationSponsorsMedia kit Overview Course overview C. elegans is a popular experimental model that combines transparency, reproducible anatomy, traceable cell lineage, sequenced genome and facile genetics, allowing to dissect animal development, physiology, and behaviour across scales of genes, cells and tissues. This 7-day course will present experimental practices in C. elegans research, while introducing historic and up-to-date findings in various research topics. Each day includes lectures, practicals, student poster presentations and interactive discussions with faculty. Various methodologies commonly employed in C. elegans research will be presented in parallel practicals with rotating student groups by relevant experts. These experimental techniques span forward and reverse genetics, genome editing, lineaging, imaging approaches...
10 June - 14 June 2024
Systems biology: from large datasets to biological insight
This course covers the use of computational tools to extract biological insight from omics datasets The content will explore a range of approaches ranging from network inference and data integration to machine learning and logic modelling that can be used to extract biological insights from varied data types Together these techniques will provide participants with a useful toolkit for designing new strategies to extract relevant information and understanding from large scale biological data The motivation for running this course is a result of advances in computer science and high performance computing that have led to groundbreaking developments in systems biology model inference With the comparable increase of publicly available large scale biological data the challenge now lies in interpreting them in a biologically valuable manner Likewise machine learning approaches are making a significant impact in our analysis of large omics datasets and the extraction of useful biological knowledge... This course covers the use of computational tools to extract biological insight from omics datasets. The content will explore a range of approaches – ranging from network inference and data integration to machine learning and logic modelling – that can be used to extract biological insights from varied data types. Together these techniques will provide participants with a useful toolkit for designing new strategies to extract relevant information and understanding from large-scale biological data.The motivation for running this course is a result of advances in computer science and high-performance computing that have led to groundbreaking developments in systems biology model inference. With the comparable increase of publicly-available, large-scale biological data, the challenge now lies in interpreting them in a biologically valuable manner. Likewise, machine learning approaches are...
17 June - 21 June 2024
Summer school in bioinformatics
Thank you to those of you who have applied we have now selected our participants and wait list There are no more spaces available Please register your interest for the 2025 course here Details of the 2024 course can be found below This course provides an introduction to the use of bioinformatics in biological research giving you guidance for using bioinformatics in your work whilst also providing hands on training in tools and resources appropriate to your research You will initially be introduced to bioinformatics theory and practice including best practices for undertaking bioinformatics analysis data management and reproducibility You will be required to review some pre recorded material for their group project prior to the start of the course Group projectsA major element of this course is a group project where you ll be placed in small groups to work together on a challenge set by trainers from EMBL EBI and external institutes This allows you to explore the bioinformatics tools and resources available in your area of interest and apply them to a set problem providing you with hands on experience relevant to your own research The group work will culminate in a presentation session involving everyone on the final day of the course giving an opportunity for wider discussion on the benefits and challenges of working with biological data Groups are mentored and supported by the trainers who set the initial challenge but the groups will be responsible for driving their projects forward with all members expected to take an active role Groups are pre organised before the course and all group members will be sent some short homework in preparation for your project work prior to the start of the course Basic outlines of the projects on offer this year are given below In your application you must indicate your first and second choice of project based on which you think would benefit your research most Not all projects may be offered and final decisions on which projects will be run during the course will be made based on the number of applicants per project Most of the projects cover mammalian data sets however in many cases the methods and approaches taught are transferable to data from various species Group project one Genome variation across human populations Natural variation between individuals or between different human populations is a result of genome mutations throughout evolutionary history Some mutations may become fixed because of their beneficial effect while most drift among individuals During this project you will investigate genomic variation between two separate human populations of European and Asian descent Using sequence data from a number of individuals from each population you will use a range of bioinformatics tools to discover variants that exist between them In the second section of the project you will attempt to analyse the functional consequences of the variants you have identified attempting to find clinical association and linking them to phenotypes Project mentor Anu Shivalikanjli EMBL EBI Group project two Interpreting functional information from large scale protein structure dataThis project will introduce you to the wealth of publicly available data in the Protein Data Bank PDB and give you the opportunity to investigate how large subsets of structure data can be used to analyse protein features and determine function In the project you will learn how to identify relevant protein structures collate and interpret functional information and implement this process programmatically Project mentor Marcus Bage EMBL EBI and Joseph Ellaway EMBL EBIGroup project three Modelling cell signalling pathwaysCurating models of biological processes is an effective training in computational systems biology where the curators gain an integrative knowledge of biological systems modelling and bioinformatics You will learn to encode and simulate ordinary differential equation models of signalling pathways from a recent publication using user friendly software such as COPASI even without extensive mathematical background You will learn to perform in silico experiments new predictions and develop hypotheses Furthermore you will learn how to annotate models and re use pre existing models from open repositories such as BioModels Project mentors Rahuman Sheriff EMBL EBI and Krishna Tiwari EMBL EBI Group project four Improving AI based bioimage analysis Artificial Intelligence AI algorithms outperform classical image analysis methods however the performance of these models is highly dependent on the quality of the annotated image datasets used to train them In this project you will explore the application of AI for biological imaging and the relationship between model training data and model performance You will use models stored in the BioImage Model Zoo and data in the BioImage Archive to fine tune and aggregate AI outputs The aim of this project will be to test evaluate and improve model performance on a diverse set of microscopy images and annotations within the BioImage Archive You will learn how to apply train tune and employ the most performant state of the art computer vision models This project serves as a valuable demonstration of how FAIR Findable Accessible Interoperable Reusable data plays an essential role in the training and enhancement of AI models Project mentors Aybuke Kupcu Yoldas EMBL EBI and Craig Russel EMBL EBI Group project five Single cell RNA sequencing analysis with PythonIn this project you will learn how to perform single cell RNA sequencing data analysis to investigate cell type heterogeneity and expression differences across conditions The analysis will be based on the SCANPY framework in Python You will start by collecting the raw count matrix and relevant metadata from the Single cell Expression Atlas After constructing the AnnData objects you will perform quality control preprocessing dimensionality reduction cell type annotation and differential expression analysis We will also explore the batch effect and its correction Project mentors Yuyao Song EMBL EBI and Anna Vathrakokoili Pournara EMBL EBI Group project six Networks and pathwaysThis project will cover typical bioinformatics analysis steps needed to put differentially expressed genes into a wider biological context You will start with gene expression data RNA seq to build an initial interaction network Next you will learn to combine public network datasets identify key regulators of biological pathways and explore biological function through network analysis You will get first hand experience in integration and co visualising with additional data and functional enrichment analysis All this helps to put the initial results into a previously known context and provide hypotheses for potential follow up experiments We will use Cytoscape Expression Atlas g Profiler StringDb among other tools We may also give a few R packages a try Project mentor Priit Adler University of Tartu... Thank you to those of you who have applied, we have now selected our participants and wait list. There are no more spaces available. Please register your interest for the 2025 course here.Details of the 2024 course can be found below.This course provides an introduction to the use of bioinformatics in biological research, giving you guidance for using bioinformatics in your work whilst also providing hands-on training in tools and resources appropriate to your research.You will initially be introduced to bioinformatics theory and practice, including best practices for undertaking bioinformatics analysis, data management, and reproducibility. You will be required to review some pre-recorded material for their group project prior to the start of the course.Group projectsA major element of this course is a group project, where you'll be placed in small groups to work together on a...
18 June - 21 June 2024
Innate immunity in host-pathogen interactions
Gordon Brown, Michaela Gack, Felix Randow, Dominique Soldati-Favre, Cyril Zipfel
OverviewSpeakersPreliminary programmePractical informationSponsorsAboutMedia kit Overview This conference will take place at EMBL Heidelberg with the option to attend virtually Symposium overview This follow up meeting to the successful EMBO EMBL Symposia Innate immunity in Host Pathogen Interactions in 2016 2018 and 2022 will provide a discussion and networking platform for all... OverviewSpeakersPreliminary programmePractical informationSponsorsAboutMedia kit Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Symposium overview This follow-up meeting to the successful EMBO | EMBL Symposia ‘Innate immunity in Host Pathogen Interactions’ in 2016, 2018 and 2022 will provide a discussion and networking platform for all scientists interested in the biology of pathogen infection. The symposium will promote the exchange of conceptual ideas in this rapidly evolving field, and also foster cross-disciplinary collaborations among the scientists. Innate immunity is studied by both microbiologists and immunologists; however, their perspectives differ widely. Microbiologists are necessarily specialists for their pathogen(s) of choice, while immunologists tend to focus on the host response. The goal of the conference is...
24 June - 28 June 2024
Cancer genomics and transcriptomics
This course will focus on the analysis of data from genomic studies of cancer It will also highlight the application of transcriptomic analysis and single cell technologies in cancer Talks and interactive sessions will give an insight into the bioinformatic concepts required to analyse such data whilst practical sessions will enable the participants to apply statistical methods to the analysis of cancer genomics data under the guidance of the trainers Virtual courseParticipants will learn via a mix of pre recorded lectures live presentations and trainer Q A sessions Practical experience will be developed through group activities and trainer led computational exercises Live sessions will be delivered using Zoom with additional support and asynchronous communication via Slack Pre recorded material may be provided before the course starts that participants will need to watch read or work through to gain the most out of the actual training event In the week before the course there will be a brief induction session Computational practicals will run on EMBL EBI s virtual training infrastructure meaning participants will not require access to a powerful computer or install complex software on their own machines Participants will need to be available between the hours of 08 45 17 00 BST each day of the course Trainers will be available to assist answer questions and provide further explanations during these times... This course will focus on the analysis of data from genomic studies of cancer. It will also highlight the application of transcriptomic analysis and single-cell technologies in cancer. Talks and interactive sessions will give an insight into the bioinformatic concepts required to analyse such data, whilst practical sessions will enable the participants to apply statistical methods to the analysis of cancer genomics data under the guidance of the trainers.Virtual courseParticipants will learn via a mix of pre-recorded lectures, live presentations, and trainer Q&A sessions. Practical experience will be developed through group activities and trainer-led computational exercises. Live sessions will be delivered using Zoom with additional support and asynchronous communication via Slack.Pre-recorded material may be provided before the course starts that participants will need to watch,...
1 July - 5 July 2024
Bioinformatics for immunologists
This course will provide participants with an introduction to a range of bioinformatics resources and approaches applicable to immunological research Participants will gain experience in analysis pipelines for NGS experiments relevant to immunology including immune receptor sequencing RNA sequencing single cell RNA sequencing and flow cytometry data analysis Additionally participants will be introduced to how data from several sources can be integrated to provide a wider view of their research thereby enabling them to be more confident users of their own data and that from public sources... This course will provide participants with an introduction to a range of bioinformatics resources and approaches applicable to immunological research. Participants will gain experience in analysis pipelines for NGS experiments relevant to immunology including immune receptor sequencing, RNA sequencing, single-cell RNA sequencing, and flow cytometry data analysis.Additionally, participants will be introduced to how data from several sources can be integrated to provide a wider view of their research, thereby enabling them to be more confident users of their own data and that from public sources.
8 July - 12 July 2024
Time-resolved macromolecular serial crystallography
Oskar Aurelius, Shibom Basu, Gisela Brändé, Arwen Pearson, Daniele de Sanctis
OverviewSpeakers and trainersPreliminary programmePractical informationSponsorsMedia kit Overview Course overview The advent of serial crystallography at free electron laser sources and the following adaptation of these methods at 3rd and 4th generation synchrotrons have opened new possibilities to perform room temperature time resolved studies in macromolecular crystallography This... OverviewSpeakers and trainersPreliminary programmePractical informationSponsorsMedia kit Overview Course overview The advent of serial crystallography at free electron laser sources and the following adaptation of these methods at 3rd and 4th generation synchrotrons have opened new possibilities to perform room temperature time resolved studies in macromolecular crystallography. This course aims to train the next generation of researchers ini) sample preparation: Participants will be trained and taught in the best practice to obtain homogenous microcrystals and in characterisation prior to loading using different sample delivery methods,ii) crystal delivery methods and data collection at a 4th generation synchrotron serial crystallography beamline: The new ID29 and similar endstations at other sources, presents experimental setup and data collection protocols that are unique in the...
8 July - 13 July 2024
Time-resolved STED nanoscopy in life sciences
Marko Lampe, Andrea Mülter, Manuel Gunkel, Julia Roberti
OverviewSpeakersPreliminary ProgrammePractical informationSponsorsMedia kit Overview Course overview This six day comprehensive advanced microscopy course focuses on stimulated emission depletion microscopy STED and its application to biological questions Through lectures tutorials and practical workshops the course provides in depth knowledge on all aspects of STED Nanoscopy It... OverviewSpeakersPreliminary ProgrammePractical informationSponsorsMedia kit Overview Course overview This six-day comprehensive advanced microscopy course focuses on stimulated emission depletion microscopy (STED) and its application to biological questions. Through lectures, tutorials and practical workshops, the course provides in-depth knowledge on all aspects of STED Nanoscopy. It covers the fundamentals of STED microscopy, different technical implementations and has a strong emphasis on sample preparation – including choice of fluorophores and refractive index matching, as well as image acquisition strategies for fixed samples and living cells. Students will have the opportunity for exercises on state-of the art STED microscopes in which they will acquire high quality nanoscopy data. The faculty consists of invited guest researchers, experts from the EMBL and Leica Microsystems....
15 July - 19 July 2024
Proteomics bioinformatics
This course provides hands on training in the basics of mass spectrometry MS and proteomics bioinformatics You will receive training on how to use search engines and post processing software quantitative approaches MS data repositories the use of public databases for protein analysis annotation of subsequent protein lists and incorporation of information from molecular interaction and pathway databases The practical elements of the course will take raw data from a proteomics experiment and analyse it You will be able to go from MS spectra to identifying and quantifying peptides and finally to obtaining lists of protein identifiers that can be analysed further using a wide range of resources The final aim is to provide you with the practical bioinformatics knowledge you need to go back to the lab and process your own data when collected This course is organised in association with the Vlaams Instituut voor Biotechnologie VIB the Flemish Institute for Biotechnology Pre recorded material may be provided before the course starts that you will need to watch read or work through to gain the most out of the actual training event In person courseThe course fee is inclusive of four nights of accommodation and catering including dinner We plan to deliver this course in person onsite at our training suite at EMBL EBI Hinxton Please be aware that we are continually evaluating the ongoing pandemic situation and as such may need to change the format of courses at short notice Your safety is paramount to us you can read our COVID guidance policy for more information All information is correct at time of publishing... This course provides hands-on training in the basics of mass spectrometry (MS) and proteomics bioinformatics. You will receive training on how to use search engines and post-processing software, quantitative approaches, MS data repositories, the use of public databases for protein analysis, annotation of subsequent protein lists, and incorporation of information from molecular interaction and pathway databases.The practical elements of the course will take raw data from a proteomics experiment and analyse it. You will be able to go from MS spectra to identifying and quantifying peptides, and finally to obtaining lists of protein identifiers that can be analysed further using a wide range of resources. The final aim is to provide you with the practical bioinformatics knowledge you need to go back to the lab and process your own data when collected.This course is organised in association...
18 August - 26 August 2024
Advances in cryo-electron microscopy and 3D image processing
Julia Mahamid, Sebastian Eustermann, Simone Mattei, Lori Passmore, Henning Stahlberg
This course provides the theoretical and practical foundations of advanced cryoEM techniques from sample preparation to data collection to image processing... OverviewSpeakersPreliminary programmePractical informationSponsors Overview Course overview Technology advances in cryo-electron microscopy (cryoEM) over the last decade have transformed our understanding of biomolecular structures across different spatial scales. CryoEM has unique capabilities for obtaining structural information in situ, within complex (cellular) environments, and to obtain atomic-level structures of biologically flexible and heterogeneous macromolecular assemblies. With the advent of the cryoEM ‘resolution revolution’, the method has undergone a tremendous growth in user base. Yet, emerging breakthroughs, such as new (correlated) imagine technologies and in AI-based image processing, demonstrate the potential for cryoEM to make another leap forward towards new frontiers. Therefore, in depth advanced training is essential to ensure future generations can...
24 August - 27 August 2024
Transcription and chromatin
Shelley Berger, Eileen Furlong, Luca Giorgetti, Jürg Müller
The meeting brings together leading experts covering all aspects of transcription including cis regulatory function long range regulation 3 dimensional looping the basal transcriptional machinery RNA polymerase regulation and function nucleosome positioning chromatin modifications chromatin remodelling and epigenetic inheritance of transcriptional silencing... OverviewSpeakersPreliminary programmePractical informationSponsorsMedia kit Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Conference overview The EMBL ‘Transcription and chromatin’ conference has a long-standing tradition in shaping the field of transcriptional regulation. The meeting brings together leading experts covering all aspects of transcription including cis-regulatory function, long range regulation, 3-dimensional looping, the basal transcriptional machinery, RNA polymerase regulation and function, nucleosome positioning, chromatin modifications, chromatin remodelling, and epigenetic inheritance of transcriptional silencing. The meeting contains many talks selected from the abstracts that are interspersed with invited speakers, discussing the latest breakthroughs in transcriptional regulation. The conference is...
1 September - 6 September 2024
Genome engineering: CRISPR/Cas in cells and mice
Neil Humphreys, Kristof Kersse, Birgit Koch, Elisabeth Zielonka
OverviewSpeakersPreliminary programmePractical informationSponsorsMedia kit Overview Course overview This course will provide training in genome editing and cell engineering in mammalian cells and mouse embryos using the highly efficient CRISPR Cas9 system Participants will learn design of CRISPR targets using bioinformatics tools generation of gene knock outs knock ins and target... OverviewSpeakersPreliminary programmePractical informationSponsorsMedia kit Overview Course overview This course will provide training in genome editing and cell engineering in mammalian cells and mouse embryos using the highly efficient CRISPR/Cas9 system. Participants will learn design of CRISPR targets using bioinformatics tools, generation of gene knock-outs/knock-ins, and target validation using the most current technologies. The course will be split into two groups (mammalian cells OR mice) based on participants’ expertise. Please note that all participants are expected to engage in all sessions. Participants should plan for approximately 1 hour extra per day for self paced learning modules. Audience This course is aimed at researchers who are familiar with basic molecular and cell biology techniques and who want to learn how to create an engineered mammalian cell...
1 September - 10 September 2024
Current methods in cell biology
Niccolò Banterle, Beth Cimini, Simone Koehler
The course aims to expose researchers with different backgrounds to the diverse cutting edge methods developed for cell biology to showcase how integrated approaches can address the current big questions in cell biology... OverviewSpeakersPreliminary programmePractical informationSponsorsMedia kit Overview Course overview In the past decade, cell biology has undergone transformational changes resulting from improvements in single molecule imaging technologies and in-situ structural biology. At the same time, the emergence of deep learning approaches that have revolutionised the field of image analysis together with techniques that complement imaging approaches to e.g. dissect the mechanical properties of cells have provided a wealth of quantitative high-dimensional data for cell biologists. The wealth of data available to cell biologists also comes with new challenges in data handling, data analysis, and data interpretation. To allow researchers to design appropriate experiments and translate the data into meaningful knowledge requires insight into which techniques are best suited to address a specific...
2 September - 5 September 2024
Imaging mouse development
Jan Ellenberg, Kate McDole, Isabel Espinosa Medina, Robert Prevedel, Shankar Srinivas
OverviewSpeakersPreliminary ProgrammePractical informationSponsorsMedia kit Overview This conference will take place at EMBL Heidelberg with the option to attend virtually Conference overview This workshop aims to bring together and help establish an emerging community with the common goal of four dimensional reconstruction of mammalian development A number of long standing questions... OverviewSpeakersPreliminary ProgrammePractical informationSponsorsMedia kit Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Conference overview This workshop aims to bring together and help establish an emerging community with the common goal of four dimensional reconstruction of mammalian development. A number of longstanding questions surrounding the early development of mammals have been inaccessible to molecular research due to technical limitations and poor physical accessibility of these processes. How is the body plan established in the early embryo? How is morphogenesis coordinated with lineage specification? The recent revolutions in advanced light microscopy and bioimage informatics have opened up completely new avenues to address these questions and make fundamental advances in our understanding of the developing mammalian...
9 September - 16 September 2024
Membrane protein expression, purification, and characterisation 3 (mPEPC3)
Maria Garcia Alai, Simon Mortensen, Kim Remans, Henning Tidow
OverviewSpeakers and TrainersProgrammePractical informationSponsorsMedia kit Overview Course overview This hands on course focuses on training participants on how to work with membrane proteins how to troubleshoot in the wet lab during solubilisation and reconstitution and how to conduct experiments at the beamlines and during early stages of cryoEM screening To perform integrated... OverviewSpeakers and TrainersProgrammePractical informationSponsorsMedia kit Overview Course overview This hands-on course focuses on training participants on how to work with membrane proteins, how to troubleshoot in the wet lab during solubilisation and reconstitution, and how to conduct experiments at the beamlines and during early stages of cryoEM screening. To perform integrated structural biology experiments well equipped crystallisation and cryo-EM facilities, as well as beamlines for SAXS for time-resolved macromolecular crystallogrophy will be used. Audience Applicants in the early stage of their research careers (mid to end of PhD project or early postdoctoral career) will be given preference based on their application and letters of reference. We also welcome applications from non-academic labs. Modules Transient transfection of mammalian cells for recombinant IMP expression...
EMBL Hamburg
9 September - 12 September 2024
Chemical biology 2024
Yimon Aye, Bryan Dickinson, Maja Köhn
OverviewSpeakersPreliminary programmePractical informationSponsorsMedia kit Overview This conference will take place at EMBL Heidelberg with the option to attend virtually Conference overview The conference sets the stage for the dissemination and exchange of new developments in diverse emerging fields within chemical biology It is the largest and longest standing conference in the... OverviewSpeakersPreliminary programmePractical informationSponsorsMedia kit Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Conference overview The conference sets the stage for the dissemination and exchange of new developments in diverse, emerging fields within chemical biology. It is the largest and longest-standing conference in the field, being a platform for inspiration, collaboration, and networking for researchers in academia and industry. We aim to provide the possibility for every chemical biologist to meet and discuss with peers topics ranging from tool development to biological applications, from computational protein design to new drug modalities. Session topics Chemical biology meets drug discovery Chemical biology of posttranslational modifications Non-coding RNA and unusual RNA products Lipids and lipidation Chemical...
15 September - 22 September 2024
Synthetic biology in action: engineering synthetic systems
Sven Panke, Vitor Pinheiro, Karen Polizzi, Nico Claassens, Velia Siciliano
OverviewSpeakersPreliminary ProgrammePractical InformationSponsorsMedia kit Overview Course overview Synthetic Biology SynBio has become a rapidly developing area for tackling a large number of challenges in life science research This is due to a combination of foundational advances in technologies such as large scale DNA synthesis and new methods that allow integrating such raw... OverviewSpeakersPreliminary ProgrammePractical InformationSponsorsMedia kit Overview Course overview Synthetic Biology (SynBio) has become a rapidly developing area for tackling a large number of challenges in life science research. This is due to a combination of foundational advances in technologies, such as large-scale DNA synthesis and new methods that allow integrating such raw synthesis power into engineering approaches. Those include genome wide small RNA regulation, dCas9-based regulation, and multiplexed genome engineering. Such fresh approaches fuel new types of biotechnological applications with immense impact on both fundamental science and society. This course will be offered in both onsite and virtual format, with different modules available to each format. It will provide the best and most exciting novelties in the field and focus on exploiting tools and concepts of SynBio...
17 September - 20 September 2024
Reconstructing the human past: using ancient and modern genomics
Johannes Krause, Ida Moltke, Maanasa Raghavan, Pontus Skoglund
Combining genome wide data from ancient and modern populations opens new windows into the past Population scale sequencing projects investigating past and present human diversity have already provided us with extraordinary insights into patterns of human variation and mobility through time and space This meeting will involve scientists from population genetics bioinformatics microbiology anthropology archaeology and history and will strengthen future interactions in this young research field that is already changing the way we think about our past and will shape how we study genetic variation in the future... OverviewSpeakersPreliminary programmePractical informationSponsorsAboutMedia kit Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Symposium overview Combining genome-wide data from ancient and modern populations opens new windows into the past and, importantly, their integration with archaeological evidence and historical records elucidates aspects of human history and cultural evolution of past societies. Population-scale sequencing projects investigating past and present human diversity have already provided us with extraordinary insights into patterns of human variation and mobility through time and space. Moreover, genome-wide data from archaic human remains, such as Neandertals and Denisovans, allows to investigate human evolution in action and to provide direct insights into genetic changes that define our own lineage. The available...
23 September - 27 September 2024
Single-cell RNA-seq analysis with Python
This course covers the analysis of single cell RNA seq data using Python and command line tools Participants will be guided through droplet based single cell RNA seq analysis pipelines from raw reads to cell clusters Furthermore you ll learn how to generate common plots for visualisation and analysis of gene expression data including TSNE UMAP and violin plots Please note that participants will not analyse their own data as part of the course There will however be ample opportunity to discuss their research and ideas with other course participants and trainers Virtual courseParticipants will learn via a mix of pre recorded lectures live presentations and trainer Q A sessions Practical experience will be developed through trainer led computational exercises and group activities Live sessions will be delivered using Zoom with additional support and asynchronous communication via Slack Pre recorded material may be provided before the course starts that participants will need to watch read or work through to gain the most out of the actual training event In the week before the course there will be a brief induction session Computational practicals will run on EMBL EBI s virtual training infrastructure meaning participants will not require access to a powerful computer or install complex software on their own machines Participants will need to be available between the hours of 09 00 17 30 BST each day of the course Trainers will be available to assist answer questions and provide further explanations during these times... This course covers the analysis of single cell RNA-seq data using Python and command line tools. Participants will be guided through droplet-based single cell RNA-seq analysis pipelines from raw reads to cell clusters. Furthermore, you'll learn how to generate common plots for visualisation and analysis of gene expression data, including TSNE, UMAP, and violin plots.Please note that participants will not analyse their own data as part of the course. There will, however, be ample opportunity to discuss their research and ideas with other course participants and trainers.Virtual courseParticipants will learn via a mix of pre-recorded lectures, live presentations, and trainer Q&A sessions. Practical experience will be developed through trainer-led computational exercises and group activities. Live sessions will be delivered using Zoom with additional support and asynchronous...
Metagenomics bioinformatics at MGnify
Learn about the tools processes and analysis approaches used in the field of genome resolved metagenomics This course will cover the use of publicly available resources to manage share analyse and interpret metagenomics data focussing primarily on assembly based approaches The delivered content will involve participants learning via live lectures and presentations followed by live Q As with the trainers Practical experience will be developed in group activities and in computational exercises run using containerised tools on our training infrastructure... Learn about the tools, processes, and analysis approaches used in the field of genome-resolved metagenomics.This course will cover the use of publicly available resources to manage, share, analyse, and interpret metagenomics data, focussing primarily on assembly-based approaches.The delivered content will involve participants learning via live lectures and presentations, followed by live Q&As with the trainers. Practical experience will be developed in group activities and in computational exercises run using containerised tools on our training infrastructure.
30 September - 3 October 2024
Defining and defeating metastasis
Eduard Batlle, Johanna Joyce, Daniel Klimmeck, Joan Massagué, Samra Turajlic
This symposium will bring together researchers from diverse fields to enhance our understanding of the dissemination and metastatic colonisation of tumour cells It will provide a unique opportunity for interdisciplinary exchange on current approaches and future collaborations on metastasis and its therapeutic challenges... OverviewSpeakersPreliminary programmePractical informationSponsorsAboutMedia kit Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Symposium overview In its second edition, this symposium will cover emerging key concepts of latent metastasis and dissemination of tumour cells including evolution and genetic diversity of metastasis, circulating tumour cells, stemness, dormancy and stromal reprogramming, transcriptional and epigenetic control, motility and invasive signalling, metastatic heterogeneity, EMT and plasticity, immune evasion, and metabolic adaptations. The meeting will provide a unique interdisciplinary exchange on current approaches and future collaborations on metastasis and its therapeutic challenges. Metastasis research is a nexus for several of the most exciting areas and technical developments of current cancer biology and...
8 October - 11 October 2024
Molecular mechanisms in evolution and ecology
Gilles Fischer, Jordi van Gestel, Kimberley Kline, Sara Mitri, Antonis Rokas
This conference series has become known for exploring yeasts as models to study questions of general relevance to evolution and ecology Experts will speak on a range of topics including evolution theory ecology genetics cell biology genomics systems biology metabolic modelling and engineering... OverviewSpeakersPreliminary programmePractical informationSponsorsMedia Kit Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Conference overview Dramatic advances in omics technologies and genetic engineering now enable us to tackle one of biology’s most fascinating problems – how evolution works in life’s communities and ecosystems. Gaining mechanistic insights requires bringing together multiple disciplines and viewpoints. The focus of this workshop series is on interspecies and cell-environment interactions across living organisms, with a special focus on bacteria, microbial eukaryotes, and viruses. The workshop series aims to highlight the latest technological advances, amplify scientific and societal focus on biodiversity, and showcase studies on the vital role of microbes in planetary ecosystems. The 2024 meeting will bring together...
13 October - 18 October 2024
Metabolite and species dynamics in microbial communities
Sunil Laxman, Kiran Patil, Elisabeth Zielonka, Maria Zimmermann-Kogadeeva
The main goal of the course is to equip the participants with the state of the art concepts in microbial ecosystems and introduce them to the key tools towards gaining mechanistic insights in community dynamics... OverviewSpeakersPreliminary programmePractical informationSponsorsMedia kit Overview This is a two-hub course taking place in parallel at both EMBL Heidelberg, Germany and the Bangalore Life Science Cluster (BLiSC) Campus in Bangalore, India. Applicants can apply for either location and are encouraged to apply for the one that is closest to them. Course overview Metagenomics studies are rapidly uncovering the compositional richness of microbial communities in diverse habitats ranging from the oceans to the human gut. While their fundamental role in our health and environment is undeniable, there is an urgent need for unravelling molecular mechanisms underlying the dynamics of these communities. This requires a combined experimental and computational approach. Recent studies (including those from the organisers’ and speakers’ labs) have underlined the success of such integrative...
EMBL Heidelberg, Other
15 October - 18 October 2024
The complex life of RNA
Elena Conti, Olivier Duss, Torben Heick Jensen, Karla Neugebauer
This symposium brings together leaders in the RNA field post docs and students with the aim of disseminating and discussing the most recent findings of information added to mRNAs The programme will include among others sessions on transcription RNA processing and modification mRNA export and localisation mRNA surveillance and decay translation and the control of mRNA expression by microRNAs... OverviewSpeakersProgrammePractical informationSponsorsAboutMedia kit Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Symposium overview Before the genetic information stored in DNA can be used to direct cell growth and metabolism, it has to be transferred into RNA. Messenger RNAs (mRNAs) that code for proteins and noncoding RNAs are key components in the transmission of genetic information in all life forms – from viruses to complex mammalian organisms. Following the great success of the 2022 edition of this long-standing symposium, we are looking forward to returning in 2024, bringing together leaders in the RNA field, post-docs and students, with the aim of disseminating and discussing the most recent results. The programme will include topics on transcription, RNA processing and modification, mRNA export and localisation, mRNA...
21 October - 26 October 2024
Imaging down to single-molecule resolution: STED & MINFLUX nanoscopy
Clara Gürth, Marko Lampe, Julia Menzel, Sebastian Schnorrenberg
OverviewSponsorsSpeakers trainers and staffMedia kit Overview Registration is not yet open for this event If you are interested in receiving more information please register your interest Course overview The aim of the course is to train microscopy users different super resolution microscopy methods Stimulated Emission Depletion STED and MINFLUX nanoscopy Both techniques overcome... OverviewSponsorsSpeakers, trainers, and staffMedia kit Overview Registration is not yet open for this event. If you are interested in receiving more information please register your interest. Course overview The aim of the course is to train microscopy users different super-resolution microscopy methods: Stimulated Emission Depletion (STED) and MINFLUX nanoscopy. Both techniques overcome the diffraction limit of light with the help of a donut shaped laser beam, and thereby achieve resolutions that are impossible with conventional light microscopy.The course combines scientific lectures by experienced researchers from the super-resolution field with hands-on sessions at the different super-resolution microscopes and in the laboratory. Participants will gain a technical understanding of the methods as well as an overview of different application fields and imaging techniques, sample...
29 October - 31 October 2024
Managing a bioinformatics core facility
This knowledge exchange workshop is an opportunity for managers of bioinformatics core facilities to learn from EMBL EBI s service teams and from each other These facilities play an essential role in enabling research in the life sciences The landscape is constantly evolving as new research tools emerge as experiments become increasingly data intensive and as their users experimental researchers become more exposed to the power of data driven biology This course will allow participants to share experiences discuss challenges and solutions that they face and plan ways to cope with the ever changing demands raised by the molecular life science field It will include sessions to learn from bioinformatics service providers hear how others have tackled common problems and work together to design core services resource them and measure their impact A limited number or travel bursaries are available Please see the Additional information tab for conditions... This knowledge exchange workshop is an opportunity for managers of bioinformatics core facilities to learn from EMBL-EBI’s service teams, and from each other. These facilities play an essential role in enabling research in the life sciences. The landscape is constantly evolving as new research tools emerge, as experiments become increasingly data intensive, and as their users – experimental researchers – become more exposed to the power of data-driven biology.This course will allow participants to share experiences, discuss challenges and solutions that they face, and plan ways to cope with the ever changing demands raised by the molecular life science field. It will include sessions to learn from bioinformatics service providers, hear how others have tackled common problems, and work together to design core services, resource them, and measure their impact.A limited number or travel...
4 November - 8 November 2024
Structural bioinformatics
This course provides a guide to the commonly used methods and tools in structural bioinformatics to analyse and interpret experimentally determined and AI predicted macromolecular structure data Structural biology determining the three dimensional shapes of biomacromolecules and their complexes can tell us a lot about how these molecules function and the roles they play within a cell Data derived from structure determination experiments and Artificial Intelligence AI assisted structure prediction enables life science researchers to address a wide variety of questions This course explores bioinformatics data resources and tools for the investigation analysis and interpretation of both experimentally determined and predicted biomacromolecular structures It will focus on how best to analyse and interpret available structural data to gain useful information given specific research contexts The course content will also cover predicting function and exploring interactions with other macromolecules This course is currently under development please check back again soon for further details... This course provides a guide to the commonly used methods and tools in structural bioinformatics to analyse and interpret experimentally determined and AI-predicted macromolecular structure data. Structural biology, determining the three-dimensional shapes of biomacromolecules and their complexes, can tell us a lot about how these molecules function and the roles they play within a cell. Data derived from structure determination experiments and Artificial Intelligence (AI)-assisted structure prediction enables life-science researchers to address a wide variety of questions. This course explores bioinformatics data resources and tools for the investigation, analysis, and interpretation of both experimentally determined and predicted biomacromolecular structures. It will focus on how best to analyse and interpret available structural data to gain useful information given...
The fundamentals of high-end cell sorting
Beata Ramasz, Diana Ordonez
OverviewSpeakersPreliminary programmePractical informationSponsorsMedia kit Overview Course overview This course includes a robust theoretical part on the fundamentals of cell sorting delivered by world leading experts and practical sessions on using standard cuvette sorters In addition the course program will include principles applications and hands on sessions on Image Cell Sorting ICS... OverviewSpeakersPreliminary programmePractical informationSponsorsMedia kit Overview Course overview This course includes a robust theoretical part on the fundamentals of cell sorting delivered by world-leading experts and practical sessions on using standard cuvette sorters. In addition, the course program will include principles, applications, and hands-on sessions on Image Cell Sorting (ICS), allowing attendees to get in touch with this exciting technology! Audience The course is intended for Ph.D. students and postdocs who are starting to use or already using cell sorting in their research projects. Applicants with previous active knowledge of basic principles of flow cytometry and regular use of flow cytometers in daily research will profit most from the course content. Modules/Resources The topics that will be covered include; instrument set-up and validation, correct operation to...
5 November - 8 November 2024
DNA replication: from basic biology to disease
Anja Groth, Thanos Halazonetis, Helle Ulrich
OverviewSpeakersProgrammePractical informationSponsorsAboutMedia Kit Overview This conference will take place at EMBL Heidelberg with the option to attend virtually Symposium overview There has been tremendous progress in the past few years regarding our understanding of DNA replication in eukaryotes both yeast and mammals Many important questions in the field are poised to be answered... OverviewSpeakersProgrammePractical informationSponsorsAboutMedia Kit Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Symposium overview There has been tremendous progress in the past few years regarding our understanding of DNA replication in eukaryotes, both yeast and mammals. Many important questions in the field are poised to be answered within the next decade. These include understanding DNA replication at the biochemical and three-dimensional protein structure levels. In addition, studies using high throughput technologies at the cellular and organismal levels are poised to answer how accurate replication of the genome is ensured by controlling origin firing in space and time. Session topics Replisome structure Origin selection, licensing and firing Chromatin replication Replication stress and fork stability Replication in cancer...
14 November - 15 November 2024
The next generation in infection biology
Maria Bernabeu, Jan Kosinski, Eva Kowalinski, John Lees, Lizette de Paula, Nassos Typas
Infectious diseases are among the most prevalent causes of human illness and death in the world Pathogens infect humans and all life forms on Earth being able to cross species barriers thereby adversely impacting human and planetary health Fundamental research in Infection Biology is urgently needed to find therapies for longstanding and emerging infectious diseases... OverviewPractical informationSponsorsMedia kit Overview Registration is not yet open for this event. If you are interested in receiving more information please register your interest. After the success of the virtual 2022 edition, this conference will return in a virtual format. Conference overview Infectious diseases are among the most prevalent causes of human illness and death in the world. Pathogens infect humans and all life forms on Earth, being able to cross species barriers, thereby adversely impacting human and planetary health. Fundamental research in infection biology is urgently needed to find therapies for longstanding and emerging infectious diseases. The EMBL Conference ‘The next generation in infection biology’ is a platform for late-stage postdoctoral scientists in the field of infection biology to present their work and connect with the leading...
17 November - 22 November 2024
Humanized mice: immunotherapy and regenerative medicine
Christian Münz, Jan Jacob Schuringa, Leonard Shultz, Brian Soper, Renata Stripecke, Anja Wege
OverviewSpeakersPreliminary programmePractical informationSponsorsMedia kit Overview Course overview Humanized mouse models became an integral part of basic and translational biomedical research for the understanding of human pathologies and for development of new therapies Human cells or tissues implanted in mice can reflect human hematopoiesis immunity oncology infections physiology... OverviewSpeakersPreliminary programmePractical informationSponsorsMedia kit Overview Course overview Humanized mouse models became an integral part of basic and translational biomedical research for the understanding of human pathologies and for development of new therapies. Human cells or tissues implanted in mice can reflect human hematopoiesis, immunity, oncology, infections, physiology and genetic polymorphisms. The development of immunotherapeutic drugs, cell and tissue therapies rely on the preclinical testing to assess their potency and safety. “Big pharma” and biotech companies include humanized mice in the non-clinical development portfolio. Humanized mouse models have varied among laboratories regarding the use of new transgenic and knock-out strains, improved handling methods, innovative sources and manipulation techniques of the human graft material and novel technological...
17 November - 24 November 2024
Integrative structural biology: solving molecular puzzles
Arjen Jakobi, Jan Kosinski, Maya Topf
This practical course offers a hands on introduction to integrative structural biology Participants will delve into the fundamentals of various experimental structure determination techniques including X ray crystallography cryo EM small angle X ray scattering NMR and mass spectrometry through both lectures and practical exercises... OverviewSpeakersProgrammePractical informationSponsorsMedia kit Overview Course overview Integrative structural biology has become a key strategy for determining the structure of biological macromolecules and their complexes. This method entails characterising the 3D structure through an array of complementary techniques, subsequently combining the data to form a consensus model using computational methodologies. This practical course offers a hands-on introduction to integrative structural biology. Participants will delve into the fundamentals of various experimental structure determination techniques including X-ray crystallography, cryo-EM, small-angle X-ray scattering, NMR, and mass spectrometry, through both lectures and practical exercises. Experts in the field will guide participants on how to analyse and interpret experimental data, and construct structural models across...
18 November - 22 November 2024
Plant genomes: from data to discovery
This course will take you through a selection of genomic tools and open resources to help you explore plant genomes Covering genomic genetic variation expression and protein data this course will show you where to find relevant data and how to use publicly available tools to connect across resources supporting plant genomic data driven knowledge discovery This course is currently under development please check back again soon for further details... This course will take you through a selection of genomic tools and open resources to help you explore plant genomes. Covering genomic, genetic variation, expression, and protein data, this course will show you where to find relevant data and how to use publicly available tools to connect across resources supporting plant genomic data-driven knowledge discovery.This course is currently under development, please check back again soon for further details.
Genome bioinformatics: from short- to long-read sequencing
A guide to the technology analysis workflows tools and resources for next generation sequencing data analysis This course will provide insights and training into how biological knowledge can be derived from genomics experiments and explain different approaches in analysing such data The main focus will be on introducing sequence informatics re sequencing differences between short and long read sequencing and variant calling during the analysis of higher eukaryotes with an emphasis on human genetic research Throughout the week more advanced topics will introduce the creation of pipelines automation and the scaling up of analysis experiments Practical sessions will be run on datasets prepared by the trainers not on personal research data Participants will learn how to process these training datasets and to apply appropriate statistical methods in their analyses This course is currently under development please check back again soon for further details... A guide to the technology, analysis workflows, tools, and resources for next-generation sequencing data analysis. This course will provide insights and training into how biological knowledge can be derived from genomics experiments and explain different approaches in analysing such data. The main focus will be on introducing sequence informatics, re-sequencing, differences between short- and long-read sequencing, and variant calling during the analysis of higher-eukaryotes, with an emphasis on human genetic research. Throughout the week, more advanced topics will introduce the creation of pipelines, automation, and the scaling-up of analysis experiments. Practical sessions will be run on datasets prepared by the trainers, not on personal research data. Participants will learn how to process these training datasets and to apply appropriate statistical methods in their analyses....
19 November - 22 November 2024
Quantitative biology to molecular mechanisms
Lacramioara Bintu, Hartland Jackson, Arnaud Krebs, Tineke Lenstra, Mikhail Savitski
OverviewSpeakersPractical informationSponsorsMedia kit Overview This conference will take place at EMBL Heidelberg with the option to attend virtually and is being organised by EMBL with generous support from the Bayer Foundation Conference overview Recent decades have witnessed a profound transformation in molecular biology research fueled by revolutionary technologies that have greatly... OverviewSpeakersPractical informationSponsorsMedia kit Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually and is being organised by EMBL with generous support from the Bayer Foundation Conference overview Recent decades have witnessed a profound transformation in molecular biology research, fueled by revolutionary technologies that have greatly enhanced our ability to systematically and quantitatively measure biological molecules. The use of genomics, proteomics, and imaging have become routine, generating vast amounts of high-resolution data that characterise biological systems across multiple scales, ranging from molecules to organisms. This technological leap allows us to describe biological systems in quantitative terms with unprecedented resolution and provides a solid foundation for formulating mechanistic models that elucidate the...
3 December - 6 December 2024
Hands-on course for sample preparation and data analysis in proteomics
Mandy Rettel, Mikhail Savitski, Frank Stein
OverviewSpeakersPreliminary programmePractical informationSponsorsMedia kit Overview Course overview The course will give an introduction to the theoretical aspects of the analysis of proteins peptides by mass spectrometry Participants will get hands on experience in sample preparation for a quantitative comparison across samples and perform a state of the art sample preparation and tandem... OverviewSpeakersPreliminary programmePractical informationSponsorsMedia kit Overview Course overview The course will give an introduction to the theoretical aspects of the analysis of proteins/peptides by mass spectrometry. Participants will get hands-on experience in sample preparation for a quantitative comparison across samples and perform a state of the art sample preparation and tandem mass tag labelling. Samples prepared by the participants will be measured by liquid chromatography-tandem mass spectrometry. In a session focused on bioinformatics analysis, participants will use FragPipe and FragPipe-Analyst in order to process the acquired data. Audience The course is intended for scientists that use or want to use proteomics for their research. Applicants should have an understanding of protein biology and should know the principle of proteomics. Ideally, they have some prior...
9 February - 14 February 2025
In situ CLEM at room temperature and in cryo
Ori Avinoam, Mandy Bormel, Rachel Mellwig, Martin Schorb
During this course we will focus on a specific method that has been developed at EMBL that enables the localisation of fluorescent proteins with high accuracy at the ultrastructural level The course will cover both the theory and practicals of high precision CLEM as well as opportunities to discuss their research with CLEM experts and receive practical and troubleshooting advice... OverviewSponsors Overview Registration is not yet open for this event. If you are interested in receiving more information please register your interest. Course overview Correlative Light and Electron Microscopy (CLEM) allows precise localisation of an event of interest within cells and tissues. During this course, we will focus on a specific method that has been developed at EMBL that enables the localisation of fluorescent proteins with high accuracy at the ultrastructural level. The course will cover both the theory and practicals of high precision CLEM, as well as opportunities to discuss their research with CLEM experts and receive practical and troubleshooting advice. Audience This course is intended for scientists with experience in EM who want to further develop their skills in high-accuracy correlated microscopy. Modules/resources The course will combine lectures and...
17 February - 21 February 2025
EMBL-EBI Training
This course covers the analysis of single cell RNA sequencing scRNA seq data using Python and command line tools Participants will be guided through droplet based scRNA seq analysis pipelines from raw reads to cell clusters You will explore and interpret single cell RNA seq data using Python as well as the Single Cell Expression Atlas Finally you will put their knowledge into practice through a group challenge on the last day Please note that you will not analyse your own data as part of the course There will however be ample opportunity to discuss your research and ideas with other course participants and trainers Virtual courseParticipants will learn via a mix of pre recorded lectures live presentations and trainer Q A sessions Practical experience will be developed through group activities and trainer led computational exercises Live sessions will be delivered using Zoom with additional support and asynchronous communication via Slack Pre recorded material may be provided before the course starts that participants will need to watch read or work through to gain the most out of the actual training event In the week before the course there will be a brief induction session Computational practicals will run on EMBL EBI s virtual training infrastructure meaning you will not require access to a powerful computer or install complex software on your own machines Participants will need to be available between the hours of 09 00 17 30 BST each day of the course Trainers will be available to assist answer questions and provide further explanations during these times This course is currently under construction and so details are subject to change Please note that we will operate this course virtually Hybrid options are not currently available We reserve the right to change the format of this course or cancel it... This course covers the analysis of single cell RNA sequencing (scRNA-seq) data using Python and command line tools. Participants will be guided through droplet-based scRNA-seq analysis pipelines from raw reads to cell clusters. You will explore and interpret single-cell RNA seq data using Python as well as the Single Cell Expression Atlas. Finally, you will put their knowledge into practice through a group challenge on the last day.Please note that you will not analyse your own data as part of the course. There will, however, be ample opportunity to discuss your research and ideas with other course participants and trainers.Virtual courseParticipants will learn via a mix of pre-recorded lectures, live presentations, and trainer Q&A sessions. Practical experience will be developed through group activities and trainer-led computational exercises. Live sessions will be delivered using...
24 February - 28 February 2025
Introduction to RNA-seq and functional interpretation
Gain an introduction to the technology data analysis tools and resources used in RNA sequencing and transcriptomics The content will provide a broad overview of the subject area and introduce participants to basic analysis of transcriptomics data using the command line It will also highlight key public data repositories and methodologies that can be used to start the biological interpretation of expression data Topics will be delivered using a mixture of lectures practical exercises and open discussions Computational work during the course will use small example data sets and there will be no opportunity to analyse personal data Virtual courseParticipants will learn via a mix of pre recorded lectures live presentations and trainer Q A sessions Practical experience will be developed through group activities and trainer led computational exercises Live sessions will be delivered using Zoom with additional support and asynchronous communication via Slack Pre recorded material may be provided before the course starts that participants will need to watch read or work through to gain the most out of the actual training event In the week before the course there will be a brief induction session Computational practicals will run on EMBL EBI s virtual training infrastructure meaning participants will not require access to a powerful computer or install complex software on their own machines Participants will need to be available between the hours of 09 00 16 00 hrs GMT each day of the course Trainers will be available to assist answer questions and provide further explanations during these times... Gain an introduction to the technology, data analysis, tools, and resources used in RNA sequencing and transcriptomics. The content will provide a broad overview of the subject area, and introduce participants to basic analysis of transcriptomics data using the command line. It will also highlight key public data repositories and methodologies that can be used to start the biological interpretation of expression data. Topics will be delivered using a mixture of lectures, practical exercises, and open discussions. Computational work during the course will use small, example data-sets; and there will be no opportunity to analyse personal data. Virtual courseParticipants will learn via a mix of pre-recorded lectures, live presentations, and trainer Q&A sessions. Practical experience will be developed through group activities and trainer-led computational exercises. Live sessions...
3 March - 7 March 2025
Introduction to multi-omics data integration and visualisation
With the increase in the volume of data across the whole spectrum of biology more opportunities as well as challenges have been created to identify novel perspectives and answer questions in life sciences This may also include public domain data which can provide added value to data derived through the researcher s own work and inform experimental design The introductory course will highlight the challenges that researchers face in integrating multi omics data sets using biological examples The course will focus on the use of public data resources and open access tools for enabling integrated working with an emphasis on data visualisation This course will not include systems biology modelling or machine learning A major element of this course is a group project where participants will be placed in small groups to work together on a challenge set by trainers from EMBL EBI data resource and research teams This allows people to explore the bioinformatics tools and resources introduced in the course and to apply these to a set problem providing hands on experience of relevance to their own research The group work will culminate in a presentation session involving all participants on the final day of the course allowing wider discussion on the benefits and challenges of integrating data... With the increase in the volume of data across the whole spectrum of biology, more opportunities, as well as challenges, have been created to identify novel perspectives and answer questions in life sciences. This may also include public domain data, which can provide added value to data derived through the researcher’s own work and inform experimental design. The introductory course will highlight the challenges that researchers face in integrating multi-omics data sets using biological examples. The course will focus on the use of public data resources and open access tools for enabling integrated working, with an emphasis on data visualisation. This course will not include systems biology modelling or machine learning.A major element of this course is a group project, where participants will be placed in small groups to work together on a challenge set by trainers from EMBL-EBI...
17 March - 21 March 2025
Gene-environment interactions in human health and disease
This introductory course explores the complex interplay between genes and the environment in shaping human phenotypes Participants will gain knowledge of cohort datasets environmental readouts and computational resources to investigate gene environment interactions Ethical and legal frameworks related to data privacy consent and genetic discrimination will be discussed The course also covers computational modelling techniques for integrating genotype and environmental effects By the end attendees will be equipped to analyse cohort datasets make use of computational resources and interpret the results Please note that you will not analyse your own data as part of the course There will however be ample opportunity to discuss your research and ideas with other course participants and trainers... This introductory course explores the complex interplay between genes and the environment in shaping human phenotypes. Participants will gain knowledge of cohort datasets, environmental readouts, and computational resources to investigate gene-environment interactions. Ethical and legal frameworks related to data privacy, consent, and genetic discrimination will be discussed. The course also covers computational modelling techniques for integrating genotype and environmental effects. By the end, attendees will be equipped to analyse cohort datasets, make use of computational resources, and interpret the results. Please note that you will not analyse your own data as part of the course. There will, however, be ample opportunity to discuss your research and ideas with other course participants and trainers
18 March - 21 March 2025
Mechanisms of drug resistance and tolerance in bacteria, fungi and cancer
Judith Berman, Markus Ralser, Luiz Carvalho, Marina Rodnina
OverviewSpeakersSponsorsAboutMedia kit Overview This conference will take place at EMBL Heidelberg with the option to attend virtually Registration is not yet open for this event If you are interested in receiving more information please register your interest Resilience to antimicrobial drugs is a rapidly increasing problem of global health Similarly cancer cells that become drug... OverviewSpeakersSponsorsAboutMedia kit Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Registration is not yet open for this event. If you are interested in receiving more information please register your interest. Resilience to antimicrobial drugs is a rapidly increasing problem of global health. Similarly, cancer cells that become drug-resistant or tolerant affect therapy outcomes, for instance during cancer chemotherapy. The underlying biological mechanisms of tolerance, resistance, and persistence are increasingly being studied and reveal a complex interplay between (cellular) evolution, biochemistry, and metabolism. Understanding these mechanisms holds the long-term promise of developing new therapeutic strategies less prone to drug resistance and, in the short term, enhances our understanding of clonal evolution and single-cell...
23 March - 28 March 2025
Measuring translational dynamics by ribosome profiling
Pasha Baranov, Jan Medenbach, Sebastian Leidel, Elisabeth Zielonka
Quantitative assessment of mRNA translation is critical to understanding gene expression in many biological settings In this practical course we will summarise the state of the art in the theoretical sessions combined with hands on experiments and computational analyses Participants will learn a robust protocol and how to adapt it to different experimental systems and also learn the key steps of computational quality control... OverviewSponsors Overview Registration is not yet open for this event. If you are interested in receiving more information please register your interest. Course overview Regulation of translation is key in the control of gene expression, playing a critical role in many biological and pathophysiological settings. In the last decade, methodological advances have provided comprehensive insights into cellular transcriptomes and proteomes, however, the systems-wide analysis of translation remained an unsolved experimental challenge. The advent of ‘ribosome-profiling’ provided the methodological basis for comprehensive transcriptome-wide analyses of translation at sub-codon resolution by the precise and high-throughput determination of ribosomal positions on mRNA. Its application in a variety of organisms yielded novel insights into protein synthesis and its dynamic regulation, significantly...
31 March - 4 April 2025
6 May - 9 May 2025
Theory and concepts in biology
Alexander Aulehla, Jordi Garcia-Ojalvo, Kirsty Wan
The goal of this symposium is to assemble an interdisciplinary community and to provide a platform to discuss perspectives on the role of theory in biology and importantly also vice versa the impact that biology can have on theory... OverviewSponsorsAbout Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Registration is not yet open for this event. If you are interested in receiving more information please register your interest. Symposium overview Recent progress in biology has led to an explosion of factual knowledge about the living world over a huge range of scales in space and time. Yet, many questions about how to make sense of this data remain across the entire diversity of the living world from the smallest bacterium to the ecosystems of the Serengeti. This meeting aims to bring together a diverse collection of researchers interested in finding ways to parlay this ever-increasing factual knowledge into a corresponding array of conceptual knowledge. The goal of this symposium is to assemble an interdisciplinary community and to provide a platform to discuss...
12 May - 16 May 2025
13 May - 16 May 2025
Chromatin and epigenetics
Asifa Akhtar, Amanda Fisher, Dirk Schubeler, Bing Ren
Epigenetics refers to heritable changes in gene expression that do not involve changes to the underlying DNA sequence At least three systems including DNA methylation histone modifications and non coding RNAs ncRNA are considered to play fundamental roles in epigenetic regulation Research over the last two decades has uncovered the role of epigenetics in a variety of human disorders and fatal diseases Moreover influence of age environment lifestyle and disease state on epigenetic states is being increasingly appreciated and actively studied This conference provides an international forum for cutting edge research in chromatin and epigenetics It provides the focal hub for people to present their research and exchange ideas in a virtual format Renowned speakers will cover the latest advances in the field... OverviewSpeakersSponsorsMedia kit Overview Registration is not yet open for this event. If you are interested in receiving more information please register your interest. This conference will take place at EMBL Heidelberg, with the option to attend virtually. Conference overview Epigenetics refers to heritable changes in gene expression that do not involve changes to the underlying DNA sequence. At least three systems including DNA methylation, histone modifications and non-coding RNAs (ncRNA) are considered to play fundamental roles in epigenetic regulation. Research over the last two decades has uncovered the role of epigenetics in a variety of human disorders and fatal diseases. Moreover influence of age, environment, lifestyle, and disease state on epigenetic states is being increasingly appreciated and actively studied. This conference provides an international forum for cutting...
18 May - 23 May 2025
This practical course will cover advanced light microscopy techniques and participants will learn how to derive qualitative and quantitative insights on molecular mechanisms in cells and developing organisms Invited guest researchers together with experts from the EMBL and microscopy professionals will foster an intense information flow with a balance of lectures and practical workshops The focus of the course will be on the use of fluorescence microscopy to obtain information about protein localisation dynamics and functions at different spatial scales... OverviewSponsorsMedia Kit Overview Registration is not yet open for this event. If you are interested in receiving more information please register your interest. Course overview This practical course will cover advanced light microscopy techniques and participants will learn how to derive qualitative and quantitative insights on molecular mechanisms in cells and developing organisms. Invited guest researchers together with microscopy experts from EMBL will foster an intense information flow with a balance of lectures and practical workshops. The focus of the course will be on the use of fluorescence microscopy to obtain information about localisation and functions of proteins and other bio-molecules at different spatial scales. Audience This course is directed towards researchers in the life sciences with access to light microscopy equipment, who have an immediate need to apply the...
20 May - 23 May 2025
26 May - 28 May 2025
Infection: pathogens, hosts and microbiomes
Maria Bernabeu, Pascale Cossart,Hiten Madhani, Kim Orth, Michael Way
OverviewSpeakersSponsors Overview This conference will take place at EMBL Heidelberg with the option to attend virtually Registration is not yet open for this event If you are interested in receiving more information please register your interest Symposium overview Infectious diseases are among the most prevalent causes of human illness and death in the world Epidemics... OverviewSpeakersSponsors Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Registration is not yet open for this event. If you are interested in receiving more information please register your interest. Symposium overview Infectious diseases are among the most prevalent causes of human illness and death in the world. Epidemics & pandemics have shaped human evolution and history, and although for a while there were considered a solved problem for the developed world, the recent past has showed us all why this is not the case. Antimicrobial resistance, global warming, human population size increase, modern lifestyles (urbanization, traveling), decrease of wild-life habitats for pathogen reservoirs all contribute to creating an ideal setting for infectious diseases becoming an even greater challenge in the future. This symposium will bring...
1 June - 6 June 2025
This course will teach the biological researchers how to analyse biological data sets using open source software Most of the analysis will be performed with docker4seq and rCASC packages which was developed to facilitate the use of computing demanding applications in the field of NGS data analysis... OverviewSponsors Overview Registration is not yet open for this event. If you are interested in receiving more information please register your interest. Course overview This course will teach the biological researchers how to analyse biological data sets using open-source software. Most of the analysis will be performed with docker4seq and rCASC packages, which was developed to facilitate the use of computing demanding applications in the field of NGS data analysis. Docker4seq and rCASC packages use docker containers that embed demanding computing tasks (e.g. short reads mapping) into isolated containers. This approach provides multiple advantages: the user does not need to install all the software on its local server, the results generated by different containers can be organised in pipelines and reproducible research is guarantee by the possibility of sharing the docker images used...
10 June - 13 June 2025
The ageing genome: from mechanisms to disease
George Garinis, Jaqueline Jacobs, Björn Schumacher, Agnel Sfeir
OverviewSponsorsAboutMedia kit Overview Registration is not yet open for this event If you are interested in receiving more information please register your interest This conference will take place at EMBL Heidelberg with the option to attend virtually Symposium overview Ageing is often considered a complex phenotype that affects multiple organs and tissues in a time dependent... OverviewSponsorsAboutMedia kit Overview Registration is not yet open for this event. If you are interested in receiving more information please register your interest. This conference will take place at EMBL Heidelberg, with the option to attend virtually. Symposium overview Ageing is often considered a complex phenotype that affects multiple organs and tissues in a time-dependent manner. This may result in the aged population consuming a myriad of medications, each aimed to treat, but not cure, the affected tissue. Recent studies have demonstrated that the “Primary Hallmarks of Ageing” are a set of five molecular mechanisms that malfunction, independent of tissue type, and drive ageing. Hence, focusing on these five hallmarks simplifies the efforts to understand ageing-associated disease. This, the second edition of this symposium will bring together an all-new, international line-up of...
16 June - 20 June 2025
This course provides an introduction to the use of bioinformatics in biological research giving you guidance for using bioinformatics in your work whilst also providing hands on training in tools and resources appropriate to your research You will initially be introduced to bioinformatics theory and practice including best practices for undertaking bioinformatics analysis data management and reproducibility You will be required to review some pre recorded material for their group project prior to the start of the course Group projectsA major element of this course is a group project where you ll be placed in small groups to work together on a challenge set by trainers from EMBL EBI and external institutes This allows you to explore the bioinformatics tools and resources available in your area of interest and apply them to a set problem providing you with hands on experience relevant to your own research The group work will culminate in a presentation session involving everyone on the final day of the course giving an opportunity for wider discussion on the benefits and challenges of working with biological data Groups are mentored and supported by the trainers who set the initial challenge but the groups will be responsible for driving their projects forward with all members expected to take an active role Groups are pre organised before the course and all group members will be sent some short homework in preparation for your project work prior to the start of the course Basic outlines of the projects on offer this year are given below In your application you must indicate your first and second choice of project based on which you think would benefit your research most Not all projects may be offered and final decisions on which projects will be run during the course will be made based on the number of applicants per project Most of the projects cover mammalian data sets however in many cases the methods and approaches taught are transferable to data from various species The projects are currently under development... This course provides an introduction to the use of bioinformatics in biological research, giving you guidance for using bioinformatics in your work whilst also providing hands-on training in tools and resources appropriate to your research.You will initially be introduced to bioinformatics theory and practice, including best practices for undertaking bioinformatics analysis, data management, and reproducibility. You will be required to review some pre-recorded material for their group project prior to the start of the course.Group projectsA major element of this course is a group project, where you'll be placed in small groups to work together on a challenge set by trainers from EMBL-EBI and external institutes. This allows you to explore the bioinformatics tools and resources available in your area of interest and apply them to a set problem, providing you with hands-on experience...
24 June - 26 June 2025
Data visualisation for biology
This course will give insights into and practical experience in data visualisation for exploring complex and multi dimensional biological data Biological and life sciences data often consists of multiple dimensions a combination of numerical values ordered data categories and time series To make sense of this data simple plots like scatterplots barcharts and boxplots are often not enough Still visualisation is critical in gaining deep insight into the data This is why many custom visualisations originated in the biological field In this course we will dive into the topic of visualisation of complex biological data and how it can be used to gain insight into and get a feel for a dataset so that targeted analyses can be defined It will be delivered through a mixture of lectures and workshop sessions and will provide hands on experience in developing visualisations The course will focus on working with omics e g genomics transcriptomics data rather than e g imaging data... This course will give insights into, and practical experience in, data visualisation for exploring complex and multi-dimensional biological data.Biological and life sciences data often consists of multiple dimensions: a combination of numerical values, ordered data, categories, and time-series. To make sense of this data, simple plots like scatterplots, barcharts, and boxplots are often not enough. Still, visualisation is critical in gaining deep insight into the data. This is why many custom visualisations originated in the biological field.In this course, we will dive into the topic of visualisation of complex biological data and how it can be used to gain insight into and get a "feel" for a dataset, so that targeted analyses can be defined. It will be delivered through a mixture of lectures and workshop sessions, and will provide hands-on experience in developing visualisations. The...
24 June - 27 June 2025
New approaches and concepts in microbiology
Pascale Cossart, Sophie Helaine, KC Huang, Michael Laub, Nassos Typas
This symposium will cover a broad range of topics in microbiology including antibiotic related research bacterial communities and microbiome cell biology regulation signalling pathogenesis and evolution Emphasis will be placed on novel approaches and technologies from systems based to single cell that drive each field or have the potential to revolutionise future research in microbiology... OverviewSponsorsMedia kitAbout Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Registration is not yet open for this event. If you are interested in receiving more information please register your interest. Symposium overview This symposium will cover a broad range of topics in microbiology, featuring diverse research on bacterial physiology, bacterial communities and microbiomes, bacterial–host and –phage interactions, and bacteria in their natural ecological contexts. Emphasis will be placed on novel approaches and technologies (from systems-based to single-cell) that drive each field or have the potential to revolutionise future research in microbiology. The meeting is the seventh of its kind, part of a successful series which started in 2013 and held biannually since, becoming a must-attend event for scientists working with...
30 June - 10 July 2025
Transitions in developing systems: concepts to methods
Matthew Benton, Jordi van Gestel, Nicoletta Petridou, Alessandra Reversi, Hanh Vu
This hands on training course will deliver the state of the art in knowledge concepts and methods utilised to study developmental transitions across biological scales through lectures discussions and application through practical sessions on model and non model systems... OverviewSponsors Overview Registration is not yet open for this event. If you are interested in receiving more information please register your interest. Course overview Developing systems at all levels of biological organisation, from molecules to ecosystems, are subject to transitions that are critical for the functional properties and evolutionary success. However, the study of transitions has proven challenging because it requires bridging biological scales and integrating diverse disciplines. Recent technological advances in quantitative biology, the acquisition of big data and machine-learning based analysis now make it possible to bridge fields. This course will bring together a diverse group of renowned scientists (speakers/trainers) who tackle these challenging yet exciting topics, including the next generation of scientists (the trainees) who plan to, or are interested in...
7 July - 11 July 2025
14 July - 18 July 2025
This course provides hands on training in the basics of mass spectrometry MS and proteomics bioinformatics You will receive training on how to use search engines and post processing software quantitative approaches MS data repositories the use of public databases for protein analysis annotation of subsequent protein lists and incorporation of information from molecular interaction and pathway databases The practical elements of the course will take raw data from a proteomics experiment and analyse it You will be able to go from MS spectra to identifying and quantifying peptides and finally to obtaining lists of protein identifiers that can be analysed further using a wide range of resources The final aim is to provide you with the practical bioinformatics knowledge you need to go back to the lab and process your own data when collected This course is organised in association with the Vlaams Instituut voor Biotechnologie VIB the Flemish Institute for Biotechnology Pre recorded material may be provided before the course starts that you will need to watch read or work through to gain the most out of the actual training event... This course provides hands-on training in the basics of mass spectrometry (MS) and proteomics bioinformatics. You will receive training on how to use search engines and post-processing software, quantitative approaches, MS data repositories, the use of public databases for protein analysis, annotation of subsequent protein lists, and incorporation of information from molecular interaction and pathway databases.The practical elements of the course will take raw data from a proteomics experiment and analyse it. You will be able to go from MS spectra to identifying and quantifying peptides, and finally to obtaining lists of protein identifiers that can be analysed further using a wide range of resources. The final aim is to provide you with the practical bioinformatics knowledge you need to go back to the lab and process your own data when collected.This course is organised in association...
3 September - 7 September 2025
Protein synthesis and translational control
Alfredo Castello, Fátima Gebauer
This conference focuses on the process of protein synthesis from the molecular and structural understanding of the translational machinery to its regulation its implications in cell and organismal biology and its alteration in disease... OverviewSponsors Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Registration is not yet open for this event. If you are interested in receiving more information please register your interest. Conference overview Mechanisms that shape cellular proteomes are critical for life. This conference focuses on the process of protein synthesis, from the molecular and structural understanding of the translational machinery to its regulation, its implications in cell and organismal biology, and its alteration in disease. The regulation of mRNA translation is critical to maintaining cell homeostasis and mediates cellular responses to environmental, physiological, and pathological cues. Given the central importance of protein synthesis, it is unsurprising that dysregulation or defects in mRNA translation are widely linked to disease, including cancer,...
9 September - 12 September 2025
Developmental metabolism: flows of energy, matter, and information
Alexander Aulehla, Heather Christofk, Heidi Lempradl, Jonathan Rodenfels
This meeting will create an intellectual space to exchange ideas across disciplines and help form a vibrant diverse community at the interphase between developmental biology molecular metabolism mathematics and physics... OverviewSponsors Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Registration is not yet open for this event. If you are interested in receiving more information please register your interest. Conference overview The fundamental question of the role of metabolism in development has seen a revival in interest. This is fueled by recent work, shedding new light onto previously unrecognised roles of metabolism. This work revealed the intricate relationship between energy flux, spatial and temporal control of metabolites, the environment and embryogenesis. This emerging view is stimulating research designed to answer fundamental questions including: How does the cellular machinery process information linked to the flow of energy and matter? How does metabolic activity impact and constrain the emergence of form and function? What are the...
6 October - 10 October 2025
Learn about the tools processes and analysis approaches used by MGnify in the field of genome resolved metagenomics This course will cover the use of publicly available resources to manage share analyse and interpret metagenomics data focussing primarily on the assembly based approaches used in MGnify analysis The delivered content will involve participants learning via live lectures and presentations followed by live Q As with the trainers Practical experience will be developed in group activities and in computational exercises run using containerised tools on our training infrastructure This course is currently under development please check back again soon for further details... Learn about the tools, processes, and analysis approaches used by MGnify in the field of genome-resolved metagenomics. This course will cover the use of publicly available resources to manage, share, analyse, and interpret metagenomics data, focussing primarily on the assembly-based approaches used in MGnify analysis. The delivered content will involve participants learning via live lectures and presentations, followed by live Q&As with the trainers. Practical experience will be developed in group activities and in computational exercises run using containerised tools on our training infrastructure. This course is currently under development, please check back again soon for further details.
22 October - 25 October 2025
Organoids: model organism development and discovery in 3D
Meritxell Huch, Karl R. Koehler, Madeline Lancaster, Esther Schnapp
The aim of this meeting is to bring together researchers from different fields to enhance our understanding of how organoids can be formed and maintained how they can be used to study disease and how we might eventually use them to regenerate and replace human organ tissue... OverviewSponsorsAbout Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Registration is not yet open for this event. If you are interested in receiving more information please register your interest. Symposium overview The ability to grow human tissues from stem cells in 3D culture has the potential to revolutionise the drug discovery process and regenerative medicine. Building on a long tradition of cell and developmental biology knowledge, organoids resembling a variety of human tissues have been generated. This conference will bring together the leading researchers in this field to establish a new research community and reveal parallels between various tissue models. The aim of this meeting is to bring together researchers from different fields to enhance our understanding of how organoids can be formed and maintained, how they can be...
27 October - 31 October 2025
28 October - 30 October 2025
The epitranscriptome
Michaela Frye, Tsutomiu Suzuki, Chengqi Yi
OverviewSponsorsMedia Kit Overview This conference will take place at EMBL Heidelberg with the option to attend virtually Event details to be confirmed pending EMBO funding application Registration is not yet open for this event If you are interested in receiving more information please register your interest Conference Overview The regulation of the transcriptome is key to... OverviewSponsorsMedia Kit Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Event details to be confirmed, pending EMBO funding application Registration is not yet open for this event. If you are interested in receiving more information please register your interest. Conference Overview The regulation of the transcriptome is key to cellular processes that underpin cell biology, development and tissue function. All classes of cellular RNA are subject to post-transcriptional modification, be it by direct chemical modification, editing or non-templated nucleotide additions. It is now emerging that the modification status of the transcriptome is dynamic and responsive to environmental/developmental cues. Together, this has elicited the realisation of an ‘epitranscriptome’ where post-transcriptional RNA modification coupled with recruitment of...
17 November - 21 November 2025
25 November - 27 November 2025
This knowledge exchange workshop is an opportunity for managers of bioinformatics core facilities to learn from EMBL EBI s service teams and from each other These facilities play an essential role in enabling research in the molecular life sciences The landscape is constantly evolving as new research tools emerge as experiments become increasingly data intensive and as their clients experimental researchers become more exposed to the power of data driven biology This course will allow participants to share experiences discuss challenges and solutions that they face and plan ways to cope with the ever changing demands raised by the molecular life science field It will include sessions to learn from bioinformatics service providers hear how others have tackled common problems and work together to design core services resource them and measure their impact This course is currently under development please check back again soon for further details... This knowledge exchange workshop is an opportunity for managers of bioinformatics core facilities to learn from EMBL-EBI’s service teams, and from each other. These facilities play an essential role in enabling research in the molecular life sciences. The landscape is constantly evolving as new research tools emerge, as experiments become increasingly data-intensive, and as their clients – experimental researchers – become more exposed to the power of data-driven biology. This course will allow participants to share experiences, discuss challenges and solutions that they face, and plan ways to cope with the ever-changing demands raised by the molecular life science field. It will include sessions to learn from bioinformatics service providers, hear how others have tackled common problems, and work together to design core services, resource them, and measure their impact. This course is...
2 December - 5 December 2025
Computational structural biology
Rommie Amaro, Charlotte Deane, Jan Kosinski, Christine Orengo, Sameer Velankar
This conference will update on the impact of AI based structure prediction provide a forum to present new methods and applications build a wider community by integrating the new scientists now massively joining the field and open a platform to discuss future directions and opportunities... OverviewSponsors Overview This conference will take place at EMBL Heidelberg, with the option to attend virtually. Registration is not yet open for this event. If you are interested in receiving more information please register your interest. Event details to be confirmed, pending EMBO funding application. Conference overview The field of computational structural biology is undergoing a revolution. AlphaFold, a program based on Artificial Intelligence (AI), has transformed the structural modelling of proteins and protein complexes by reaching accuracy similar to experimental structures. Over 200 million structural models have already been released in AlphaFold Database. Following AlphaFold, other protein modelling programs proliferated from researchers worldwide. AI has started advancing RNA modeling too. The wealth of protein models has sparked an explosion of new tools for structural...
No matching events found; Reset your filters .
For past events see the archive .

Sign up for our quarterly events newsletter, event email announcements and our blog.

EMBL Course and Conference Programme
The EMBL Course and Conference Programme welcomes 7000 participants annually and covers practical courses, workshops and conferences across all the six EMBL sites. Learn more.
Self-paced learning
anywhere, anytime for everyone
Learn on research carried out and technologies used at EMBL and online training on bioinformatics. Free to access anytime and anywhere.

EMBL is Europe’s leading life sciences laboratory
EMBL seeks to better understand the molecular basis of life. In the process, we’re changing European science.

IMAGES
VIDEO
COMMENTS
The EMBL International PhD Programme (EIPP), originally established in 1983, provides PhD students with an excellent starting platform for a successful career in science by fostering early independence and interdisciplinary research.. EMBL is dedicated to promoting excellence in the molecular life sciences throughout Europe.To achieve this goal, we inspire and train talented young scientists ...
Research. EMBL International PhD Programme. Established in 1983, the programme provides students with the best starting platform for a successful career in science.
EMBL PhD students receive theoretical and practical training and conduct a research project under the supervision of an EMBL faculty member, monitored by a thesis advisory committee. The duration of PhD studies is normally three-and-a-half to four years. In Year 1 all new PhD students will attend the EMBL Predoctoral Core Course in Molecular ...
As part of the Nordic EMBL Partnership for Molecular Medicine, outstanding doctoral researchers are recruited to the institute-level rotation programme through the annual call. The recruited doctoral researchers work with 2-3 different research groups during a 6-9 month period before matching to a research group in which they remain for their complete PhD studies.
Downloads This modal can be closed by pressing the Escape key or activating the close button.
The EMBL International PhD Programme receives far more applications from very good candidates than we can invite for interview. Applicants who are not short-listed, and have indicated that we can share their application, become part of our shared applicant pool. Applications will be added into the pool twice a year - in December and June.
The fully-funded EMBL International PhD Programme (EIPP) is designed to promote interdisciplinary research, international collaboration and early independence. By completing your PhD education at one of EMBL's six sites (in Barcelona, Grenoble, Hamburg, Heidelberg, Hinxton and Rome), you will develop the skills needed to excel in your future ...
EMBL PhD students at EMBL-EBI are members of the University of Cambridge and one of its Colleges. They receive their degree from Cambridge University; the programme is coordinated in Heidelberg with local support at EBI. Please visit the EMBL International PhD Programme pages to learn about how to apply. Please note all applicants must secure a ...
Many PhD students and postdocs aspire to a permanent research position at a university or research institute, but competition for such positions has increased. Here, we report a time-resolved analysis of the career paths of 2284 researchers who completed a PhD or a postdoc at the European Molecular Biology Laboratory (EMBL) between 1997 and 2020.
At EMBL, Giuseppe enjoyed the vibrant mix of languages, cultures and people. "You feel really immersed in Europe at EMBL, and the friendships that you build during your PhD stay with you long after you leave the lab," he says. As a PhD student at EMBL, Giuseppe felt encouraged to pursue his intellectual curiosity and push boundaries.
As part of the Nordic EMBL Partnership for Molecular Medicine, we welcome applications for the 2024 FIMM-EMBL International PhD Program.Detailed information regarding the position descriptions, research areas, qualifications, and application and selection process can be found at the University of Helsinki Open Positions page.. We currently have two to four openings for doctoral researchers who ...
The EMBL PhD Symposium has been arranged by EMBL PhD students with the aim of creating an occasion for connecting young researchers and high-profile scientists. The focus of this year's Symposium is on the topic "The Power of Many: Collective Behaviour Across Scales".
EMBL provides PhD students with a starting platform for a successful career in science by fostering early independence and interdisciplinary research. The enriching encounter of different nationalities, the friendly and collaborative atmosphere, and the passion for science is what unites EMBL's diverse staff and provides an ideal setting to ...
EMBL Australia gives the nation's best PhD students the opportunity to develop international networks and alliances in Europe via EMBL's programs, workshops and conferences. We also provide training programs for PhD students in Australia, giving them a head start in their science careers. Travel Grants. PhD Course. Postgraduate Symposium.
The EMBL Documents repository holds the digital copies of official EMBL documents, reports, brochures and various other publications. May 15, 2019 EMBL International PhD Programme
Many PhD students and postdocs aspire to a permanent research position at a university or research institute, but competition for such positions has increased. Here, we report a time-resolved analysis of the career paths of 2284 researchers who completed a PhD or a postdoc at the European Molecular Biology Laboratory (EMBL) between 1997 and 2020.
You have to enable javascript in your browser to use an application built with Vaadin. You have to enable javascript in your browser to use an application built with ...
EMBL International PhD Programme: Join the unique and world-renowned EMBL International PhD Programme, where exceptional candidates are invited to pursue cutting-edge research in various scientific areas across multiple EMBL sites. This exciting opportunity offers a comprehensive training experience that fosters scientific excellence and ...
The EMBL Fellows' Complementary Skills and the EMBL Fellows' Career Service provide additional career development support tailored for EMBL PhD students and postdocs. Its external training activities include a rich programme of courses and conferences, as well as supporting scientific visitors coming to the lab.
Scholarship Description: EMBL provides a starting platform for a successful scientific career through early independence and interdisciplinary research. The program offers fully funded PhD positions with comprehensive health care and pension benefits. Eligibility: Applicants with diverse backgrounds in Biology, Chemistry, Physics, Mathematics, Computer Science, Engineering, and Molecular ...
The EMBL Lautenschläger summer school, held 4-15 Jul 2022, introduced students from a range of scientific backgrounds to interdisciplinary life science research. ... "I would definitely love to pursue my PhD at this laboratory and will be looking forward to applying once I complete my current degree course." ...
EMBL delivers world-class courses, conferences and workshops at the forefront of molecular life science and its applications. EMBL Events. ... Audience Applicants in the early stage of their research careers (mid to end of PhD project or early postdoctoral career) will be given preference based on their application and letters of reference. We ...