- Search by keyword
- Search by citation
Page 1 of 12

Glutaminolysis regulates endometrial fibrosis in intrauterine adhesion via modulating mitochondrial function
Endometrial fibrosis, a significant characteristic of intrauterine adhesion (IUA), is caused by the excessive differentiation and activation of endometrial stromal cells (ESCs). Glutaminolysis is the metabolic...
- View Full Text
The long-chain flavodoxin FldX1 improves the biodegradation of 4-hydroxyphenylacetate and 3-hydroxyphenylacetate and counteracts the oxidative stress associated to aromatic catabolism in Paraburkholderia xenovorans
Bacterial aromatic degradation may cause oxidative stress. The long-chain flavodoxin FldX1 of Paraburkholderia xenovorans LB400 counteracts reactive oxygen species (ROS). The aim of this study was to evaluate the...
MicroRNA-148b secreted by bovine oviductal extracellular vesicles enhance embryo quality through BPM/TGF-beta pathway
Extracellular vesicles (EVs) and their cargoes, including MicroRNAs (miRNAs) play a crucial role in cell-to-cell communication. We previously demonstrated the upregulation of bta-mir-148b in EVs from oviductal...
YME1L-mediated mitophagy protects renal tubular cells against cellular senescence under diabetic conditions
The senescence of renal tubular epithelial cells (RTECs) is crucial in the progression of diabetic kidney disease (DKD). Accumulating evidence suggests a close association between insufficient mitophagy and RT...
Effects of latroeggtoxin-VI on dopamine and α-synuclein in PC12 cells and the implications for Parkinson’s disease
Parkinson’s disease (PD) is characterized by death of dopaminergic neurons leading to dopamine deficiency, excessive α-synuclein facilitating Lewy body formation, etc. Latroeggtoxin-VI (LETX-VI), a proteinaceo...
Glial-restricted progenitor cells: a cure for diseased brain?
The central nervous system (CNS) is home to neuronal and glial cells. Traditionally, glia was disregarded as just the structural support across the brain and spinal cord, in striking contrast to neurons, alway...
Carbapenem-resistant hypervirulent ST23 Klebsiella pneumoniae with a highly transmissible dual-carbapenemase plasmid in Chile
The convergence of hypervirulence and carbapenem resistance in the bacterial pathogen Klebsiella pneumoniae represents a critical global health concern. Hypervirulent K. pneumoniae (hvKp) strains, frequently from...
Endometrial mesenchymal stromal/stem cells improve regeneration of injured endometrium in mice
The monthly regeneration of human endometrial tissue is maintained by the presence of human endometrial mesenchymal stromal/stem cells (eMSC), a cell population co-expressing the perivascular markers CD140b an...
Embryo development is impaired by sperm mitochondrial-derived ROS
Basal energetic metabolism in sperm, particularly oxidative phosphorylation, is known to condition not only their oocyte fertilising ability, but also the subsequent embryo development. While the molecular pat...
Fibroblasts inhibit osteogenesis by regulating nuclear-cytoplasmic shuttling of YAP in mesenchymal stem cells and secreting DKK1
Fibrous scars frequently form at the sites of bone nonunion when attempts to repair bone fractures have failed. However, the detailed mechanism by which fibroblasts, which are the main components of fibrous sc...
MSC-derived exosomes protect auditory hair cells from neomycin-induced damage via autophagy regulation
Sensorineural hearing loss (SNHL) poses a major threat to both physical and mental health; however, there is still a lack of effective drugs to treat the disease. Recently, novel biological therapies, such as ...
Alpha-synuclein dynamics bridge Type-I Interferon response and SARS-CoV-2 replication in peripheral cells
Increasing evidence suggests a double-faceted role of alpha-synuclein (α-syn) following infection by a variety of viruses, including SARS-CoV-2. Although α-syn accumulation is known to contribute to cell toxic...
Lactadherin immunoblockade in small extracellular vesicles inhibits sEV-mediated increase of pro-metastatic capacities
Tumor-derived small extracellular vesicles (sEVs) can promote tumorigenic and metastatic capacities in less aggressive recipient cells mainly through the biomolecules in their cargo. However, despite recent ad...
Integration of ATAC-seq and RNA-seq identifies MX1-mediated AP-1 transcriptional regulation as a therapeutic target for Down syndrome
Growing evidence has suggested that Type I Interferon (I-IFN) plays a potential role in the pathogenesis of Down Syndrome (DS). This work investigates the underlying function of MX1, an effector gene of I-IFN,...
The novel roles of YULINK in the migration, proliferation and glycolysis of pulmonary arterial smooth muscle cells: implications for pulmonary arterial hypertension
Abnormal remodeling of the pulmonary vasculature, characterized by the proliferation and migration of pulmonary arterial smooth muscle cells (PASMCs) along with dysregulated glycolysis, is a pathognomonic feat...
Electroacupuncture promotes neurogenesis in the dentate gyrus and improves pattern separation in an early Alzheimer's disease mouse model
Impaired pattern separation occurs in the early stage of Alzheimer’s disease (AD), and hippocampal dentate gyrus (DG) neurogenesis participates in pattern separation. Here, we investigated whether spatial memo...
Role of SYVN1 in the control of airway remodeling in asthma protection by promoting SIRT2 ubiquitination and degradation
Asthma is a heterogenous disease that characterized by airway remodeling. SYVN1 (Synoviolin 1) acts as an E3 ligase to mediate the suppression of endoplasmic reticulum (ER) stress through ubiquitination and de...
Advances towards the use of gastrointestinal tumor patient-derived organoids as a therapeutic decision-making tool
In December 2022 the US Food and Drug Administration (FDA) removed the requirement that drugs in development must undergo animal testing before clinical evaluation, a declaration that now demands the establish...
Melatonin alleviates pyroptosis by regulating the SIRT3/FOXO3α/ROS axis and interacting with apoptosis in Atherosclerosis progression
Atherosclerosis (AS), a significant contributor to cardiovascular disease (CVD), is steadily rising with the aging of the global population. Pyroptosis and apoptosis, both caspase-mediated cell death mechanism...
Prenatal ethanol exposure and changes in fetal neuroendocrine metabolic programming
Prenatal ethanol exposure (PEE) (mainly through maternal alcohol consumption) has become widespread. However, studies suggest that it can cause intrauterine growth retardation (IUGR) and multi-organ developmen...
Autologous non-invasively derived stem cells mitochondria transfer shows therapeutic advantages in human embryo quality rescue
The decline in the quantity and quality of mitochondria are closely associated with infertility, particularly in advanced maternal age. Transferring autologous mitochondria into the oocytes of infertile female...
Development of synthetic modulator enabling long-term propagation and neurogenesis of human embryonic stem cell-derived neural progenitor cells
Neural progenitor cells (NPCs) are essential for in vitro drug screening and cell-based therapies for brain-related disorders, necessitating well-defined and reproducible culture systems. Current strategies em...
Heat-responsive microRNAs participate in regulating the pollen fertility stability of CMS-D2 restorer line under high-temperature stress
Anther development and pollen fertility of cytoplasmic male sterility (CMS) conditioned by Gossypium harknessii cytoplasm (CMS-D2) restorer lines are susceptible to continuous high-temperature (HT) stress in sum...
Chemogenetic inhibition of NTS astrocytes normalizes cardiac autonomic control and ameliorate hypertension during chronic intermittent hypoxia
Obstructive sleep apnea (OSA) is characterized by recurrent episodes of chronic intermittent hypoxia (CIH), which has been linked to the development of sympathoexcitation and hypertension. Furthermore, it has ...
SARS-CoV-2 spike protein S1 activates Cx43 hemichannels and disturbs intracellular Ca 2+ dynamics
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes the ongoing coronavirus disease 2019 (COVID-19). An aspect of high uncertainty is whether the SARS-CoV-2 per se or the systemic inflammation ...
The effect of zofenopril on the cardiovascular system of spontaneously hypertensive rats treated with the ACE2 inhibitor MLN-4760
Angiotensin converting enzyme 2 (ACE2) plays a crucial role in the infection cycle of SARS-CoV-2 responsible for formation of COVID-19 pandemic. In the cardiovascular system, the virus enters the cells by bind...
Two murine models of sepsis: immunopathological differences between the sexes—possible role of TGFβ1 in female resistance to endotoxemia
Endotoxic shock (ExSh) and cecal ligature and puncture (CLP) are models that induce sepsis. In this work, we investigated early immunologic and histopathologic changes induced by ExSh or CLP models in female a...
An intracellular, non-oxidative factor activates in vitro chromatin fragmentation in pig sperm
In vitro incubation of epididymal and vas deferens sperm with Mn 2+ induces Sperm Chromatin Fragmentation (SCF), a mechanism that causes double-stranded breaks in toroid-linker regions (TLRs). Whether this mechani...
Focal ischemic stroke modifies microglia-derived exosomal miRNAs: potential role of mir-212-5p in neuronal protection and functional recovery
Ischemic stroke is a severe type of stroke with high disability and mortality rates. In recent years, microglial exosome-derived miRNAs have been shown to be promising candidates for the treatment of ischemic ...
S -Nitrosylation in endothelial cells contributes to tumor cell adhesion and extravasation during breast cancer metastasis
Nitric oxide is produced by different nitric oxide synthases isoforms. NO activates two signaling pathways, one dependent on soluble guanylate cyclase and protein kinase G, and other where NO post-translationa...
Identifying pyroptosis- and inflammation-related genes in intracranial aneurysms based on bioinformatics analysis
Intracranial aneurysm (IA) is the most common cerebrovascular disease, and subarachnoid hemorrhage caused by its rupture can seriously impede nerve function. Pyroptosis is an inflammatory mode of cell death wh...
Drosophila Atlastin regulates synaptic vesicle mobilization independent of bone morphogenetic protein signaling
The endoplasmic reticulum (ER) contacts endosomes in all parts of a motor neuron, including the axon and presynaptic terminal, to move structural proteins, proteins that send signals, and lipids over long dist...
Mucin1 induced trophoblast dysfunction in gestational diabetes mellitus via Wnt/β-catenin pathway
To elucidate the role of Mucin1 (MUC1) in the trophoblast function (glucose uptake and apoptosis) of gestational diabetes mellitus (GDM) women through the Wnt/β-catenin pathway.
Human umbilical cord mesenchymal stem cells (hUC-MSCs) alleviate paclitaxel-induced spermatogenesis defects and maintain male fertility
Chemotherapeutic drugs can cause reproductive damage by affecting sperm quality and other aspects of male fertility. Stem cells are thought to alleviate the damage caused by chemotherapy drugs and to play role...
Exploring the Neandertal legacy of pancreatic ductal adenocarcinoma risk in Eurasians
The genomes of present-day non-Africans are composed of 1–3% of Neandertal-derived DNA as a consequence of admixture events between Neandertals and anatomically modern humans about 50–60 thousand years ago. Ne...
Identification and analysis of key hypoxia- and immune-related genes in hypertrophic cardiomyopathy
Hypertrophic cardiomyopathy (HCM), an autosomal dominant genetic disease, is the main cause of sudden death in adolescents and athletes globally. Hypoxia and immune factors have been revealed to be related to ...

How do prolonged anchorage-free lifetimes strengthen non-small-cell lung cancer cells to evade anoikis? – A link with altered cellular metabolomics
Malignant cells adopt anoikis resistance to survive anchorage-free stresses and initiate cancer metastasis. It is still unknown how varying periods of anchorage loss contribute to anoikis resistance, cell migr...
Single nucleotide polymorphisms associated with wine fermentation and adaptation to nitrogen limitation in wild and domesticated yeast strains
For more than 20 years, Saccharomyces cerevisiae has served as a model organism for genetic studies and molecular biology, as well as a platform for biotechnology (e.g., wine production). One of the important eco...
Investigating the dark-side of the genome: a barrier to human disease variant discovery?
The human genome contains regions that cannot be adequately assembled or aligned using next generation short-read sequencing technologies. More than 2500 genes are known contain such ‘dark’ regions. In this st...
Hyperbaric oxygen treatment increases intestinal stem cell proliferation through the mTORC1/S6K1 signaling pathway in Mus musculus
Hyperbaric oxygen treatment (HBOT) has been reported to modulate the proliferation of neural and mesenchymal stem cell populations, but the molecular mechanisms underlying these effects are not completely unde...
Polar microalgae extracts protect human HaCaT keratinocytes from damaging stimuli and ameliorate psoriatic skin inflammation in mice
Polar microalgae contain unique compounds that enable them to adapt to extreme environments. As the skin barrier is our first line of defense against external threats, polar microalgae extracts may possess res...
Correction: Utility of melatonin in mitigating ionizing radiation‑induced testis injury through synergistic interdependence of its biological properties
The original article was published in Biological Research 2022 55 :33
Beyond energy provider: multifunction of lipid droplets in embryonic development
Since the discovery, lipid droplets (LDs) have been recognized to be sites of cellular energy reserves, providing energy when necessary to sustain cellular life activities. Many studies have reported large num...
Retraction Note: Tridax procumbens flavonoids: a prospective bioactive compound increased osteoblast differentiation and trabecular bone formation
Electroacupuncture protective effects after cerebral ischemia are mediated through mir-219a inhibition.
Electroacupuncture (EA) is a complementary and alternative therapy which has shown protective effects on vascular cognitive impairment (VCI). However, the underlying mechanisms are not entirely understood.
Topsoil and subsoil bacterial community assemblies across different drainage conditions in a mountain environment
High mountainous environments are of particular interest as they play an essential role for life and human societies, while being environments which are highly vulnerable to climate change and land use intensi...
Functional defects in hiPSCs-derived cardiomyocytes from patients with a PLEKHM2-mutation associated with dilated cardiomyopathy and left ventricular non-compaction
Dilated cardiomyopathy (DCM) is a primary myocardial disease, leading to heart failure and excessive risk of sudden cardiac death with rather poorly understood pathophysiology. In 2015, Parvari's group ident...
Human VDAC pseudogenes: an emerging role for VDAC1P8 pseudogene in acute myeloid leukemia
Voltage-dependent anion selective channels (VDACs) are the most abundant mitochondrial outer membrane proteins, encoded in mammals by three genes, VDAC1 , 2 and 3 , mostly ubiquitously expressed. As 'mitochondrial ...
ABCA1 transporter promotes the motility of human melanoma cells by modulating their plasma membrane organization
Melanoma is one of the most aggressive and deadliest skin tumor. Cholesterol content in melanoma cells is elevated, and a portion of it accumulates into lipid rafts. Therefore, the plasma membrane cholesterol ...
Acupuncture regulates the apoptosis of ovarian granulosa cells in polycystic ovarian syndrome-related abnormal follicular development through LncMEG3-mediated inhibition of miR-21-3p
The main features of polycystic ovary syndrome (PCOS) are abnormal follicular development and ovulatory dysfunction, which are caused by excessive apoptosis of ovarian granulosa cells. Acupuncture has been sho...
- Editorial Board
- Manuscript editing services
- Instructions for Editors
- Sign up for article alerts and news from this journal
- Follow us on Twitter
- Follow us on Facebook
- ISSN: 0717-6287 (electronic)
Biological Research
ISSN: 0717-6287
- Submission enquiries: Access here and click Contact Us
- General enquiries: [email protected]
March 25, 2024
Potassium and fish fins
Potassium channels regulate the size of fish fins, but how? Xiaowen Jiang, Kun Zhao, Yi Sun, Christopher Antos and colleagues show that the scale of zebrafish embryonic pectoral fins is determined by the regulation of intracellular K + by retinoic acid via Rcan2 and the potassium channel Kcnk5b.
Image credit: Xiaowen Jiang & Christopher Antos
PLOS Biologue
Community blog for plos biology, plos genetics and plos computational biology..
Update Article
Wolbachia effects in mosquitoes
Wolbachia -induced cytoplasmic incompatibility in fruit flies is known to cause embryonic lethality by modifying chromatin integrity in developing sperm. Rupinder Kaur, Seth Bordenstein and co-workers reveal an analogous mechanism in the w Mel-transinfected Aedes aegypti mosquitoes that are used to control vector-borne diseases.
Image credit: Rupinder Kaur
Recently Published Articles
- Shortcut citations in the methods section: Frequency, problems, and strategies for responsible reuse
- MYCN drives oncogenesis by cooperating with the histone methyltransferase G9a and the WDR5 adaptor to orchestrate global gene transcription
- Wolbachia -bearing Aedes aegypti mosquitoes deployed for arbovirus control">The mechanism of cytoplasmic incompatibility is conserved in Wolbachia -bearing Aedes aegypti mosquitoes deployed for arbovirus control
Current Issue February 2024
Research Article
How MYCN drives neuroblastoma
The oncoprotein MYCN drives oncogenesis in neuroblastoma by activating genes involved in ribosome biogenesis and protein synthesis, while repressing neuronal differentiation, but the underlying mechanisms are unknown. Zhihui Liu, Carol Thiele and co-authors show that MYCN cooperates with the histone methyltransferase G9a and the WDR5 adaptor to orchestrate global gene transcription.
Image credit: pbio.3002240
SMARCAL1 ubiquitylation and replication fork stability
The E3 ubiquitin ligase RFDW3 regulates replication fork progression and stability, but how does it achieve this? Maïlyn Yates, Alexandre Maréchal and co-workers reveal that SMARCAL1 is a substrate of RFWD3 and that its ubiquitylation disengages it from RPA, thereby limiting its activity at stalled replication forks and preventing fork collapse.
Image credit: pbio.3002552
Methods and Resources
Tracking galactose
Galactose is an important sugar that can fuel glycolysis, but observing its metabolic dynamics in vivo has been difficult to date. Uğurcan Sakizli, Tomomi Takano and Sa Kan Yoo develop a genetically encoded sensor to measure intracellular galactose dynamics in flies.
Image credit: Ugurcan Sakizli
Syncing brains with your leader
In-group social bonding stabilizes hierarchical structures in various species. Jun Ni, Jiaxin Yang and Yina Ma show that social bonding in humans selectively increases information exchange and prefrontal neural synchronization for human leader-follower pairs.
Image credit: pbio.3002545
Peptides against SARS-CoV-2
The envelope protein is highly conserved among human coronaviruses, suggesting it could be a good drug target. Ramsey Bekdash, Kazushige Yoshida, Masayuki Yazawa and co-workers develop a platform for the identification and optimization of peptides targeting the coronavirus envelope, and show that iPep-SARS2-E inhibits SARS-CoV-2 infection in lung organoids and in mice.
Image credit: pbio.3002522
Perspective
Lysosome diversity
Single-organelle resolution approaches have the potential to advance our knowledge of the heterogeneity of lysosome function. Claudio Bussi and Maximiliano Gutierrez propose a 'lysosome states' concept that links single lysosomes to function.
Image credit: pbio.3002576
Sustainable coral reef restoration
Coral reef restoration faces many challenges. This Essay advocates prioritizing environmental and climatic considerations to increase the sustainability of future coral reefs and open up opportunities for new restoration approaches.
Image credit: pbio.3002542
Formal Comment
Ghost lineages and HGT
A recent study questioned the use of branch length methods to assess relative timing of horizontal gene transfers because of the effects of so-called “ghost” lineages. This Formal Comment discusses key considerations regarding these effects. See also this response .
Image credit: pbio.3002460
Evolutionary safety of mutagenic drugs
Some drugs increase the mutation rate of their target pathogen, raising the concern that they might thereby accelerate adaptation. This Perspective article proposes a four-step process to evaluate the evolutionary safety of such treatments.
Image credit: Yitzhak Pilpel via DALL-E
PLOS Biology 20th Anniversary
PLOS Biology is 20 and we are celebrating with a collection that contains articles that look back at landmark studies that we published, others that look past and future, and others discussing how publishing and open science have evolved and what is to come.
Engineering plants for a changing climate
This collection explores engineering strategies to help us adapt plants to a changing climate, including breeding techniques, genome engineering, synthetic biology and microbiome engineering.
Going for green
The green collection explores biological solutions that could be applied to reduce CO2 emissions, get rid of non-degradable plastics, produce food in a sustainable manner or generate energy.
Ocean solutions for a sustainable, healthy and inclusive future
This collection explores potential solutions to mitigate the impacts of human activity on ocean ecosystems to minimize or reverse degradation.
Mendel's legacy
To mark 200 years since the birth of Gregor Mendel, this collection looks back at the impact of his original research and asks how well it stands up to modern views on genetics.
EMBL: Diversity of plants: from genomes to metabolism
April 9 - 12
Meet Senior Editor Roli Roberts ([email protected])
Titisee: Organelle communication: from fundamental understanding to therapeutics
April 10 - 14
Meet Senior Editor Ines Alvarez-Garcia ([email protected])
Symposium on the Immune System of Bacteria
April 16 - 18
Meet Editor in Chief Nonia Pariente ([email protected])
RS: Vaccines and antimicrobial resistance: from science to policy
April 29 - 30
Meet Magazine Editor Joanna Clarke ([email protected])
Publish with PLOS
Submit Your Manuscript
Connect with Us
- PLOS Biology on Twitter
- PLOS on Facebook
Get new content from PLOS Biology in your inbox
Thank you you have successfully subscribed to the plos biology newsletter., sorry, an error occurred while sending your subscription. please try again later..
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Life (Basel)

Artificial Intelligence in Biological Sciences
Associated data.
Not applicable.
Artificial intelligence (AI), currently a cutting-edge concept, has the potential to improve the quality of life of human beings. The fields of AI and biological research are becoming more intertwined, and methods for extracting and applying the information stored in live organisms are constantly being refined. As the field of AI matures with more trained algorithms, the potential of its application in epidemiology, the study of host–pathogen interactions and drug designing widens. AI is now being applied in several fields of drug discovery, customized medicine, gene editing, radiography, image processing and medication management. More precise diagnosis and cost-effective treatment will be possible in the near future due to the application of AI-based technologies. In the field of agriculture, farmers have reduced waste, increased output and decreased the amount of time it takes to bring their goods to market due to the application of advanced AI-based approaches. Moreover, with the use of AI through machine learning (ML) and deep-learning-based smart programs, one can modify the metabolic pathways of living systems to obtain the best possible outputs with the minimal inputs. Such efforts can improve the industrial strains of microbial species to maximize the yield in the bio-based industrial setup. This article summarizes the potentials of AI and their application to several fields of biology, such as medicine, agriculture, and bio-based industry.
1. Introduction
There is no precise definition of artificial intelligence (AI) so far, but in general it refers to the ability of any machines which can simulate the intelligences of higher organisms. The field of AI has important roots in almost every branch of research including philosophy, mathematics, computing, psychology and biology [ 1 ]. An ideal AI system would be self-aware, logical and able to learn from experience. It would also be able to perceive and react to external environments. With the aid of algorithms based on machine learning (ML) and deep learning (DL) approaches, such an intelligent system could be developed to carry out activities that require human intellect [ 2 ]. John McCarthy in the 1956 first coined the term “artificial intelligence (AI) for an intelligent machine system at the Dartmouth Conference [ 2 ]. The earliest significant work in AI includes the contribution of mathematician Alan Mathison Turing. He proposed his ideas in a public lecture in London about the concept of self-learning and self-instructed machines that learn from their own experiences as a human being does [ 3 , 4 ]. Due to his initial observation and conceptualization of facts about smart machines, Alan Turing is widely regarded as the father of AI and modern computer science. He was an early proponent of the theory that the human brain functions essentially like a digital computer [ 5 ]. He pioneered the experiment known as “The Turing Test”, which became a pivotal moment in the development of AI ( Figure 1 ). His paper titled ‘Computing Machinery and Intelligence,’ looked into the possibility of a non-living computer thinking like a human and was a landmark in this area [ 3 ]. Several other additional significant events paved the way for the development of the AI we see today ( Figure 2 ). An AI program written by Arthur Samuel in 1952 for the IBM 701 prototype and a ‘virtual rat’ trained to move through a predefined path based on a neural network by John Holland were such groundbreaking preliminary works [ 6 , 7 ]. In 1973, a group of Japanese engineers created the first humanoid robot, which had several distinct capabilities for a machine at the time, including the ability to walk upright, hold objects and converse in Japanese.

Alan Turing designed the Turing Test in 1950. This test includes three participants, a human interrogator, an intelligent machine and another human who we can call A, B and C, respectively. A is not aware of the identity of B and C, and A can send and receive response in only the form of text messages from B and C. A may ask B and C, a variety of questions, and based on their response, if A is unable to distinguish which one of B and C is a computer, then computer B may be considered as intelligent with thinking ability. If a human interrogator A could not distinguish the difference between another human and a computer, then the computer must be intelligent enough to be considered human. This test simply is to figure out whether or not a machine has ability to think.

Timelines highlighting the important breakthroughs in the evolutionary path of artificial intelligence and its application in various fields.
Another significant event in the AI timeline was the construction of IBM’s supercomputer, Deep Blue, which was capable of playing chess completely indistinguishable from humans. It was the first artificial intelligence to defeat Grandmaster Garry Kasparov in a timed match [ 8 ] ( Figure 2 ). Successful use of AI planning and perception approaches may be seen in NASA’s space-based autonomous vehicles, which use technology to steer and move on their own without human intervention [ 4 ]. DL and ML are crucial elements of AI that train themselves by picking up knowledge from data of various sources that are either generated directly or indirectly from the natural intelligence system. The more these deep learning and machine learning algorithms are trained using data from various sources, the more advanced, intelligent and self-aware artificial systems may be developed ( Figure 3 ) [ 9 ].

Schematic representation of major components of artificial intelligence and the continuous learning process with the help of natural intelligence to make smarter machines.
AI may be classified into two broad categories, Narrow or Weak AI and Artificial General Intelligence or Strong AI. Weak AI makes some attempt to copy or mimic human cognitional thought; it enables the automation of the majority of tasks in ways that humans are incapable of [ 10 ]. The most visible examples of weak AI on a daily basis include Tesla’s autopilot feature, facial recognition on our smartphones, Google’s search engine, Instagram’s AI for understanding user interests, Apple’s Siri and Amazon’s Alexa. Strong AI is a far more advanced and complex notion than weak AI. Strong AI is not restricted by human-made laws, and it thinks and controls the system entirely on its own. In layman’s terms, it is a program or a machine that simulates precise human cognitive or intellectual qualities, such as emotions or strong problem-solving abilities [ 11 , 12 ]. A weak AI program is designed to accomplish only one task at a time; a strong AI can efficiently perform numerous tasks simultaneously [ 13 ]. Although, self-awareness is the most essential and unique quality that distinguishes strong AI from weak AI, it is still in the early stages of development, and there are no real-life applications we can observe [ 13 ].
There are techniques used in AI which include a lot of variations, for example, the rule-based systems that are based on symbolic representations and work on inferences. AI systems have an ANN-based system which is designed to work on the interface with other neurons and connection weights [ 14 ]. Despite all of these, they all share four characteristics. Firstly, they have the feature of knowledge representation. Rule-based systems, frame-based systems and semantic networks use a series of if–then rules, whereas artificial neural networks use connections and connection weights [ 15 ]. Second, AI engineered systems are capable of learning. As self-learning entities [ 16 ], they gather data, such as by choosing the appropriate connection weights for an artificial neural network or defining the rules for a rule-based expert system. Third, they have the rules which can be implicit or explicit in an AI system. The fourth is the search, which can be incorporated into the system in several ways. For instance, it can be used to find the states that lead to a solution more quickly or to find the best set of connection weights for an ANN by minimizing the fitness function [ 1 ]. Depending on the algorithm employed, AI can also be divided into “rule-based”, also known as AI in general terms, and “non-rule-based”, also known as ML. In rule-based algorithms, conditioned branching and instructions are provided in order to obtain the best solution. For instance, the algorithm would be completely true to the instruction and merge the numbers when the case is defined as, “when subject numbers of two different datasets are the same, they should be treated as duplicates and need to be merged”. A rule-based algorithm works well when there are few options available. However, the development of a rule-based algorithm is quite challenging in complex scenarios. ML, on the other hand, develops rules directly from established training input and implements them in the ML algorithms via statistical methods. Thus, ML focuses on quickly recognizing patterns from a huge volume of information to provide findings that are more reliable than manual analysis and predictions [ 17 ].
AI has now made its way into the biological field, demonstrating its worth through innovative and cutting-edge procedures [ 18 ]. Additionally, the world has seen a true revolution in the field of information technology (IT), leading to the production and storage of an enormous amount of data, not just in the field of technology but in other areas as well in recent years. Both information technology and biology have flourished during the past half-century. According to Moore’s law, the number of transistors on a chip will double about once every two years. It is a consequence of and driver for the rapid growth of information technology [ 19 ]. Computational resources are inextricably linked to big data, which encompasses annotated and raw information due to the ever-increasing volume and complexity of data from multiple sources [ 20 , 21 ]. Because of developments in sequencing and other high-throughput techniques, the biosciences and biotech industries have made remarkable strides in recent years [ 22 ]. AI-based algorithms have the capacity to effectively store and process large amounts of raw, unstructured data and make them available for quick extraction, which is necessary to build an intelligent computing system with complex decision-making capability [ 23 , 24 ]. Such advancement in data generation, storage and analysis allows the development of a wide range of products and services in different sectors including biosciences [ 19 ]. While advances in computing and the Internet ushered in the third industrial revolution and laid the groundwork for AI’s meteoric rise, Big Data and the analytics it spawned have allowed us to take our intelligence to new heights [ 25 ]. AI is now considered a major invention of the fourth industrial revolution [ 26 ]. Experiments that would have taken years to execute are now feasible and often inexpensive due to recent advances in data and methodology. Raw data in a variety of formats are generated as a result of these experimental analyses. The ability to store and analyze data with the help of AI has created new possibilities for the academic community, scientific researchers and the biotech industry. Various applications of AI are used in biology, including the precise identification of the 3D geometry of biological molecules such as proteins which is one of the most critical tasks and useful in biological research. Moreover, in biological science, AI plays a critical role in promoting innovation not only in laboratories, but also throughout the lifecycle of a medication or chemical product [ 27 ]. Furthermore, AI-based tools and applications help automate complicated production procedures, thereby meeting the fast-rising demand for medications, chemicals for use in industry and food and other bio-based raw materials. ML, a subset of AI, aids in the prediction of outcomes by executing massive permutations and combinations of datasets available for the drug molecules to determine the best combination without relying on traditional manual methods in the lab [ 28 ]. Although traditional model-driven methods are still useful for analyzing biological data, they lack the ability to use vast amounts of available data, or even big data, to uncover information, forecast data behavior and comprehend complex data linkages. The extensive use of big data is becoming increasingly important in biotechnology and bioinformatics as it continues to grow and becomes available to academicians and scientists for analysis throughout the world [ 29 ]. These data are quantified in terms of multi-omics, such as genomics, transcriptomics, proteomics, and metabolomics, from different biological sources and need to be properly annotated and analyzed to understand complex biological systems. AI and deep neural network designs might efficiently analyze genomic data to determine the genetic basis of a trait and to uncover genetic markers linked with certain traits [ 30 , 31 ]. The use of AI may aid in deciphering complex links across diverse information hidden in data to obtain meaningful insights from them. As a result, the incorporation of AI approaches is now widely observed in the field of biological science and is expected to increase further in the near future as this technology matures [ 2 ]. Furthermore, medical images and drug responses contribute complex but significant data and need efficient algorithmic programs to analyze them. Therefore, ML- and DL-based AI is garnering much attention due to their capabilities for faster processing of huge data and extraction of meaningful information. AI-based digital image processing, drug designing and virtual drug tests might transform medical science in the near future [ 32 , 33 ].
The current review article highlights how Artificial Intelligence, and its components could be used in the medical, agricultural, and bio-based industrial sectors to make human life more sustainable.
2. AI in Medical Science
Medical science and biotechnology advancements have opened new avenues for developing medications and antibiotics. AI has enormous potential for widespread applications in the pharmaceutical industry ( Figure 4 ). With AI, novel therapeutic molecules based on known target structures can be discovered [ 34 ]. A branch of AI known as ML is commonly employed in disease diagnosis since it leverages the outcomes of diagnostic testing to improve the accuracy of results [ 35 ]. AI allows researchers to manage challenging issues, including quantitative and predictive epidemiology, precision-based medicines and host–pathogen interactions [ 36 ]. AI can help in disease detection and diagnosis and make computer code more accessible to non-technical individuals [ 37 ]. Predictive epidemiology, individual-based precision medicine and the analysis of host–pathogen interactions are examples of research areas that could benefit from machine and deep learning breakthroughs [ 38 ]. These approaches aid with disease diagnosis and individual case identification, more accurate forecasts and fewer mistakes, faster decision making and better risk analysis ( Figure 4 ). The growing number of tissue biomarkers and the complexity of their evaluations significantly promote the use of AI-based techniques. These AI-based biomarkers help physicians in the prediction and analysis of the diagnosis, patient responses to the treatment and patient survival [ 39 ]. More realistic models of complex socio-biological systems are achievable because of knowledge representation and reasoning modelling [ 40 ]. ML-based methods can also be used to improve the efficiency and reliability of epidemiological models [ 41 , 42 ]. ML advances helped develop ten cellular parameters algorithmic program-based models that can accurately distinguish benign from malignant tumors [ 43 ].

A conceptual model that illustrates the application possibilities of artificial intelligence in the disciplines of health, agriculture, animal science and industrial biotechnology.
It is important to take into account individual differences in genetics, ecology and lifestyle in precision medicine [ 44 ]. Medical practitioners recognize that the metabolic, physical, physiological and genetic makeup of an individual affects how their body responds to drugs in a certain way. Despite this, we are currently employing an umbrella approach that treats all patients, regardless of their varying conditions, with the same drug. However, due in large part to advances in AI, a new era of personalized medicine, in which pharmaceuticals are tailored to the body’s needs and adaptability, is evolving. Although the transition appears to be simple, it entails a significant amount of data collection, processing, maintenance and execution [ 45 ]. Moreover, millions of prediction analyses will be included in the process to identify the best therapeutic candidate molecules for a particular case. Using this strategy, physicians and clinicians may better predict which disease treatment and preventative strategies will be most effective for particular patient groups ( Figure 4 ). Researchers could use AI in DNA, RNA and protein studies to better visualize the effects of drug doses on living tissue over time and reorganize signaling networks during therapy [ 46 , 47 ]. Based on AI, IBM Watson assists in the creation of the appropriate treatment plan for a patient depending on the patient’s medical history and personal data, including genetic makeup [ 48 ]. An AI-based system of personalized medicine will not only reduce treatment cost but also minimize the side effects of drugs in the patient [ 49 ]. In addition to saving time and improving patient care, AI can also simplify gene editing, radiography and drug management planning procedure [ 50 ]. Furthermore, electronic health records (EHRs) can be improved with evidence-based clinical decision support systems [ 44 , 51 , 52 ]. AI involves massive processing capacity (supercomputers), algorithms that can learn at a phenomenal rate (deep learning) and a new strategy that utilizes physicians’ cognitive talents ( Table 1 ). This technique can contribute to the development of innovative theoretical models of disease pathophysiology and can help forecast major adverse effects of prolonged medications [ 53 ]. In a recent study, an AI-based approach was found to be very beneficial for the early identification, diagnosis, prognosis and treatment of myopia [ 54 ]. In cardiology, dermatology and oncology, deep learning algorithms outperform physicians at least in the diagnosis of disease [ 55 , 56 , 57 ]. Evidently, computer algorithms can detect metastatic breast cancer in sentinel lymph node biopsies in full slide images with an accuracy rate of more than 91 percent, and this was raised to 99.5 percent when physician inputs were added [ 58 ]. One of the proven applications of AI in risk analysis is for diagnosing heart malfunctioning through cardiovascular imaging. It includes automated monitoring of any deviations from normal conditions based on image processing, myocardial function and the detection and analysis of coronary atherosclerotic plaques [ 59 ]. The YOLOV3 algorithm was used for AI-based medical image segmentation for 3D printing and naked eye 3D visualization to detect the prostate in T2-weighted MRI images (AIMIS3D) [ 59 ]. There are several variables that might be efficiently analyzed through AI, such as determining which conditions are resistant to certain antibiotics and not to others [ 60 ]. Such analysis can support physicians and significantly decrease unnecessary testing and costs in medical care.
AI in disease detection and prediction modelling.
It is important to underline the importance of combining these algorithms with medical expertise ( Table 1 ). New pharmaceutical compounds can be discovered via data analysis using AI, which reduces the need for clinical trials, allowing medications to be brought to market more quickly without compromising their safety [ 32 ]. Moreover, we may be able to forecast the onset of genetically predisposed diseases considerably earlier with the help of AI [ 50 ]. Patients will also be able to prevent and treat certain inherited diseases.
One of the applications of AI in the pharmaceutical industry is “Open Targets”, which is a relatively new strategic effort to explore the relationship between drug targets and diseases, as well as how certain genes are linked to diseases [ 67 ]. SPIDER is another AI technique that is being designed to determine the role of natural products in drug discovery [ 68 ]. Furthermore, quantitative structure–activity relationship (QSAR) studies are particularly useful in creating novel effective medications in a very short period of time using a computer simulation tool [ 69 ]. A QSAR model based on a radial basis function (RBF) artificial neural network (ANN) model that was trained using particle swarm optimization (PSO) technique was used in a recent study to predict the pKa values of 74 different types of drugs [ 69 ]. Natural language processing (NLP), ML, and robotic process automation are clearly the three key areas of advancement for AI in the field of medicine [ 70 , 71 ]. Natural Language Processing has recently been used to enhance colonoscopy analysis, improving accurate detection of adenoma and polyps [ 72 ]. Additionally, an ML approach may be used to predict diseases such as atrial fibrillation and urinary tract infections in certain patient groups by using models such as support vector machine (SVM) based on clinical features of the disease [ 73 , 74 , 75 ]. Similar initiatives have been utilized to improve heart disease prognosis using a heart-murmur-detecting technology [ 76 ]. The FDA has already approved up to 29 AI-based medical devices and algorithms in various fields of medical sciences [ 77 ].
The first AI-based model approved by the FDA in the healthcare sector was a diagnostic model based on an autonomous AI system, IDx-DR. This model was successfully used in to detect diabetic retinopathy with sensitivity, specificity and imageability of 87.2%, 90.7% and 96.1%, respectively, in a sample size of 819 subjects over 10 primary care units in the United States. The model was trained with a diversified sample dataset consisting of individuals of different ages, races and sex, thus minimizing the chances of errors in different groups [ 78 ]. Several randomized clinical trials (RCTs) have also been performed to test the efficacy and safety of AI and ML models in clinical practice. In an RCT (Registration number: ChiCTR-DDD-17012221), the impact of a deep-leaning-based automated polyp identification algorithm on polyp detection accuracy and adenoma detection rates (ADRs) was evaluated. In this RCT, successive patients were randomly assigned to go through colonoscopy either with or without the help of the automated polyp identification model that provided a simultaneous optical notification and sound alert upon polyp discovery. Results obtained from patients who have undergone the automated AI-based detection system outperformed the control cohorts of ADR and the average amount of adenoma and polyps detected per coloscopy. This automated technology can thus be pertinent in treatment regimens and routine practices for improved identification of colon polyps due to its great sensitivity, high precision and stable outcomes [ 79 ]. The introduction of AI systems in medical decision making has also resulted in the cost-effectiveness of complete medical treatment. In a study, the use of a procalcitonin-based decision algorithm (PCTDA) for hospitalized sepsis and lower respiratory tract infection patients led to a shortened duration of stay, lowered antibiotic administration, lesser artificial ventilation periods and decreased number of patients with infections and antibiotic resistance. On average, PCTDA-based treatment brought about a 49% and 23% decrease in overall expenses from conventional treatment for sepsis and lower respiratory tract infections, respectively [ 80 ]. The pharmaceutical industry will better grasp genetic information with improved AI and ML skills ( Figure 4 ). Evidently, when integrated with ML and NLP, robotic process automation has significant applications and has the potential to reshape medical science in the near future [ 81 ]. Despite the tremendous advancements we have observed, there is still a lot of work to be done before AI-based therapy becomes a reality.
3. AI in Agricultural Biotechnology
Face recognition [ 82 ], cancer prediction in tissue [ 83 ] and metabolic flux analysis [ 84 ] are just a few examples of significant advances made with AI approaches, and there is a potential to achieve a similar revolution in the agricultural field. According to a report published by the United Nations’ Food and Agriculture Organization (FAO), the world’s population will reach more than 9 billion by 2050 [ 85 ]. Population expansion will eventually put a strain on the agriculture sector’s ability to provide food. In order to feed the world’s growing population and advance the nation’s economy, agriculture is essential [ 86 ]. It is a significant source of revenue for a number of countries, including India.
Agriculture occupies around 38% of the planet’s total land surface [ 85 ]. The majority of agricultural activities are now manual, and agriculture may significantly benefit from automation in terms of obtained yield and invested inputs. The implementation of technological breakthroughs in agriculture may contribute to the change in rural economies and villagers’ livelihoods [ 87 , 88 ]. Agricultural techniques are generally designed to overcome a variety of obstacles, including pest infestation, inefficient use of pesticides and fertilizers, weeds, drought and a lack of an adequate irrigation system, inefficient harvesting, storage and finally marketing. The agricultural sector could be transformed by AI intervention in the areas of soil management, water requirement assessment, precise mapping of fertilizer need, pesticide, insecticide, herbicide need, yield prediction and overall crop management ( Figure 5 ) [ 89 , 90 , 91 ]. With the advancement of AI-based technology, drones and robots are being used to improve real-time monitoring of crops, harvesting and subsequent processing [ 92 ]. AI and ML techniques are currently being used by biotechnology companies to design and train autonomous robots capable of performing key agricultural activities such as crop harvesting at a much faster rate than traditional methods [ 89 ]. The data collected by drones are processed and evaluated using deep learning and computer vision techniques [ 93 ]. Machine learning approaches assist in the access and forecast of a wide range of environmental variables that influence agricultural output, such as weather fluctuations and the arrival of the monsoon in India [ 89 , 94 , 95 ]. As mentioned elsewhere, AI-based solutions in the agricultural industry help to improve efficiency and control numerous aspects such as crop yield, soil profile, crop irrigation, content sensing, weeding and crop monitoring ( Figure 5 ) [ 89 , 96 ].

Agricultural fields where artificial intelligence (AI) could have a positive impact.
Traditional and older morphological characteristic inspection is time-consuming, error-prone and costly. The machine vision method might be easily applied in agricultural practices, which can speed up and simplify the procedure while being more precise and accurate [ 93 ]. Identification and selection of improved varieties may speed up and make the process easier by using automated non-invasive, rapid scoring of various plant features through high-throughput phenotyping methods [ 97 ]. Due to the tools of AI and IoT, swarm intelligence and drone technology can now be employed for several agricultural activities [ 98 ]. Recent developments in DL- and ML-based algorithm design to estimate the price of agricultural products may enable farmers to receive a higher return on their labor and investment [ 99 ]. For effective irrigation, artificial neural networks, fuzzy logic and meta-heuristic algorithms have recently been developed [ 100 , 101 ]. According to a recent study, convolutional neural network (CNN), which takes into account several environmental variables, is one of the most trustworthy ML algorithms to estimate soybean and maize yields [ 102 ]. Recent advances in AI-based biosensors for early disease detection in crop plants, even in asymptomatic plants, have the potential to greatly minimize product loss caused by biotic stressors [ 103 ]. AI-based drone technologies such as EfficientNetV2, which are designed to detect and classify plant diseases with accuracy and precision of 99.99% and 99.63%, respectively, are one of the promising automated technologies for the monitoring of plant health in a time-saving and cost-effective manner [ 104 ]. For the detection of bacterial spot disease in plants, a hybrid AI model based on convolutional autoencoder (CAE) and CNN has also achieved 99.35% and 99.38% in the training and testing periods, respectively, [ 105 ].
The use of AI may make it simpler to identify potential targets in big genome data for genetic manipulation and design effective synthetic promoters in efforts to improve agronomic traits in plants [ 106 , 107 ]. The growing necessities for smart agriculture have resulted in substantial advancements in the area of AI-based agricultural forecasting and prediction, which has improved crop productivity to a great extent [ 93 ]. A similar attempt was made in a recent study where image datasets were analyzed by employing AI algorithms, namely ANN and genetic algorithm (GA)-based platforms, for the prediction of crop yield in an optimized manner [ 108 ]. During the training period, the model obtained a maximum validation accuracy of 98.19%, whereas a maximum accuracy of 97.75% was yielded during the test period [ 108 ]. This model worked effectively under limited resource restrictions and less data, producing optimal results [ 108 ]. In another significant study, a new methodology for predicting agricultural yield in greenhouse crops employing recurrent neural network (RNN) and temporal convolutional network (TCN) algorithms was proposed [ 109 ]. Based on previous environmental and production data, this approach can be utilized to estimate greenhouse crop yields more accurately than its standard ML and deep learning peers [ 109 ].
Furthermore, this experimental investigation has also demonstrated the crucial importance of previous yield datasets in correctly predicting future crop productivity [ 109 , 110 ]. Several million individuals in developing nations have benefited from the green revolution by preventing and combining high-yield crops, synthetic fertilizers and water. However, owing to widespread misuse of herbicides, pesticides and fertilizers, the green revolution could not be considered fully “green”. Certain approaches for high-yielding crops typically need a large amount of agro-chemicals and water [ 111 ]. AI-based approaches are being developed to reduce the reliance on noxious agro-chemicals and to attain a state of sustainability in agriculture [ 79 ]. For optimizing agricultural resources, a remote sensing assisted control system (RSCS) has been developed [ 112 ]. This methodology makes use of AL and ML technology to improve environmental sustainability while fostering novel agricultural product development planning. When analyzed with other techniques, the findings revealed that the RSCS demonstrated the highest precision, performance, data transfer rate, productivity, irrigation management and carbon dioxide release ratio of 95.1, 96.35, 92.3, 94.2, 94.7 and 21.5%, respectively, [ 112 ]. Thus, AI models have the potential to manage agricultural products and productivity in a “green” manner. In another study, an AI and machine vision-based smart sprayer was developed to spray herbicides specifically to weed targets, thus reducing weedicide overuse and environmental contamination. This sophisticated technology combined a cutting-edge weed detection concept, a unique rapid and precise spraying method and a weed mapping model with 71% and 78% precision and recall, respectively, [ 113 ]. Due to limited collecting techniques and a lack of integration of diverse data sources, data gathering from agricultural regions linked to soil hydration, crop quality or insect infestations frequently depend on manual analysis.
Meanwhile, as the industry becomes more digital, the combination of remote sensing for computerized screening and analytical techniques with datasets for soil studies, weather predictions, etc., and sophisticated AI models is reducing the need for agrochemicals [ 93 ]. In this regard, the substantial NaLamKI action plan that seeks to create AI-based open access software that could greatly help the agricultural industry has received funding from the German government. This plan seeks to develop datasets by combining information from different sensors in order to optimize different farming practices with the help of AI and ML technologies [ 93 , 114 ]. Similar governmental initiatives are required in large numbers to make farmers adapt AI on a greater scale.
In agriculture, integrating precise image-based features with omics data may aid in finding critical traits involved in stress tolerance and acclimatization mechanisms [ 115 ], as well as contribute to the development of climate resilient cropss. Farmers will be able to generate more output with less input, increase the quality of their output and ensure a faster time to market for their harvested crops owing to AI-based technology adaptation [ 93 ] ( Table 2 ). Although first-generation AI can be employed in the surveyance and classification of omics data, it is tailored for the handling of specific problems related to single-omics datasets without integrating data from other modalities [ 93 , 116 ]. In agricultural biotechnology, next-generation AI is fundamentally envisioned to dynamically ameliorate and handle large multi-omics datasets in addition to predicting the breeding value of complex traits across different environmental conditions [ 116 ].
Recently developed AI-based algorithms in the agricultural sector.
4. AI and Industrial Biotechnology
Industrial biotechnology, sometimes known as white biotechnology, is the modern application of biotechnology to the sustainable processing and manufacturing of commodities, chemicals and fuels from renewable sources using live cells and their enzymes. The demand for industrial chemicals, medicines, food-grade chemicals and other biochemistry-related raw materials has increased dramatically over the previous decade [ 121 ]. ML and AI-based technologies may aid in the design of novel pharmaceuticals and the identification of their efficacy and adverse effects before their actual production, drastically reducing the time spent bringing a drug from the lab to the market for ordinary people [ 32 ]. Microorganisms and plant/animal cells are used in biotechnological processing to make products in a variety of sectors, including drugs, pharmaceuticals, food and feed, disinfectants, pulp and textiles. In order to detect outages, optimize machinery for efficient manufacture and improve product quality, the Internet of things, ML and AI could be used effectively [ 122 ]. AI-based computer models are becoming increasingly widespread, and robotics and machine learning could be used to develop the best optimum growth conditions for the strains, as well as the degree to which valuable products can be obtained ( Figure 4 ). For instance, AI or response surface methodologies (RSM) -based approaches have been used in the high-level production of amylases from Rhizopus microsporous, using various agro-industrial wastes for optimal experimentation designs [ 123 ]. Similarly, AI algorithms such as artificial neural networks (ANN) and genetic algorithms (GA) have been integrated for the optimization of fermentation media to produce glucansucrase from Leuconostoc dextranicum . A 6% rise in glucansucrase activity was predicted by the integrated ANN-GA model over a regression-based prediction approach [ 124 ]. The application of the integrated ANN-GA model for the optimization of cellulase production by Trichoderma stromaticum under solid-state fermentation has been reported recently, and a 31.58-fold increase in cellulase production was achieved after optimization with the AI model [ 125 ].
AI-based technologies have also been used to scale up and optimize bioprocesses for enzyme production on pilot scales. A low-cost method for increasing the synthesis of extracellular laccase from Staphylococcus arlettae utilizing tea waste was performed in a study. RSM and ANN coupled with GA were two consecutive statistical methods that were employed to increase enzyme production and resulted in a sixteen times rise in enzyme yield. Moreover, a pilot scale bioprocess was established utilizing the ideal parameters identified by GA, namely tea waste (2.5%) NaCl (4.95 mM), L-DOPA (5.65 mM) and 37℃ temperature, which improved the enzyme production by 72 times [ 126 ]. Furthermore, some AI models based on the fuzzy expert system are also capable of monitoring wastewater treatment plants on a pilot scale [ 127 ].
Biofuel is one of the most important bioproducts for which the industrial production process can be enhanced using ML and AI for maximum output. In the bioenergy sector, AI-based approaches have been used to predict biomass feedstock properties, bioenergy end-uses, and bioenergy supply chains and have developed an integrated ANN-Taguchi method model for the prediction and maximization of biofuel production via torrefaction and pyrolysis [ 128 , 129 ]. Optimization and design of experimental factors were performed using the Taguchi method which led to the attainment of maximum biofuel yield up to 99.42%, whereas ANN showed linear regression prediction of 0.9999 for biochar and 0.9998 for bio-oils.
Integrated ANN-GA models have been used in the modeling and optimization of the methanolysis process of waste peanut shells for the generation of biofuels. Biofuel yield optimized by the RSM model was 16.49%, whereas that of the ANN-GA model was reported to be 17.61%. This shows that integrated ANN-GA has better optimization potential than the RSM model alone [ 130 ]. ML-based bioprocess models have also been constructed with the help of AI-based methods such as ANN, CNN, (long short-term memory networks) LSTMs, kNNs (k-nearest neighbors) and RF (random forests) for predicting the accumulation of carbohydrates in cyanobacteria biomass cultivated in wastewater for biofuel production. The finest results for approximation of system dynamics were achieved with a 1D-CNN with a mean square error of 0.0028 [ 131 ]. Textiles, new chemicals and biodegradable biopolymer synthesis could all benefit from similar processes [ 132 ]. Furthermore, it may be used to assist in the development of synthesis techniques for such biochemicals that produce the highest yield with the least amount of input ( Figure 4 ). Additionally, AI could assist in real-time forecasting of market demand for medications or chemicals. AI and ML have also helped in the production of metabolites. Systems metabolic engineering is a process that helps in the rapid production of high-performing microbial strains for the long-term production of chemicals and minerals. The increasing availability of bio big data, such as omics data, has resulted in an application for ML techniques across various stages of systems metabolic engineering, such as host strain selection, metabolic pathway reconstruction, metabolic flux optimization and fermentation [ 19 ]. Various machine learning algorithms, including deep learning, have facilitated in optimizing the bioprocess parameters and exploring a larger metabolic space that is linked to the biosynthesis of a target bioproduct [ 133 ]. This trend is also influencing biotechnology businesses to adopt ML techniques more frequently in the creation of their production systems and platform technologies [ 134 ]. In the brewery industry, AI has demonstrated promising potential to overcome fundamental shortcomings and enhance production through knowledge accumulation and automated control. In a study, AI models were constructed using aroma profiles and spectroscopic data obtained from commercial alcohol for assessing the quality traits and aroma of beer. The intelligent models resulted in highly accurate predictions for six major beer aromas [ 135 ]. Smart e-nose technologies based on ANN models have also been developed to assess the presence of different chemicals such as ethanol, methane, carbon monoxide, hydrogen sulfide, ammonia, and so forth in beer [ 136 ]. A study was involved in the development of a computer program that simulated the operation of a highly customizable three-layer feed-forward multilayer perception neural network, which using data from prior experiments, could forecast changes in the parameters of white wine alcoholic fermentation. This work provided a befitting approach for the digitalization of brewing processes, thus enabling it to be acclimatized to other intelligent and knowledge-based frameworks [ 137 ]. Another study led to the development of an innovative knowledge-based approach for controlling the batch fermentation of alcohol employed in making white wine. The primary sources of information used in developing the AI model were different case studies and experimental results, as well as the knowledge obtained from brewery experts regarding different parameters related to optimization and control of the overall process. Using the monitoring, regulation and data acquisition software of the fermentation bioreactor, an application for automated process control was developed [ 138 ]. The further incorporation of control systems, processes and innovative advancements can be greatly facilitated by such kinds of AI models, thus supporting sustainable development.
5. Challenges and limitations
Despite their immense potential, AI-based technologies have yet to make their way into everyday practice. AI models can improve the accessibility of various biological sectors; however, they may also exacerbate pre-existing discrepancies. Since AI models are extremely reliant on the datasets on which they are developed as well as the labels connected with them, prejudices against the underrepresented in the learning algorithms might be reinforced [ 139 ]. Several factors must be considered to properly assess the resilience of some deep neural networks. For the development of AI models, metadata must be created, retrieved and cleansed. Programs should further be designed and evaluated under the oversight of field professionals for analysis and correction of inaccuracies committed in practice [ 140 ]. In spite of significant advances in the design of AI and ML-based models in recent years, few have been incorporated into healthcare, and many prospects for adopting these models for everyday usage remain untapped. CNNs, for instance, were initially used in study designs commencing in 2015, primarily on dental radiographs, with the first clinical uses for these tools only recently emerging [ 141 ]. Unavailability and inaccessibility of clinical data due to organizational policies, insufficient reproducibility in processing datasets and assessing outcomes and residual concerns around accountability and transparency to patients remain the most common hurdles in adapting AI in routine medical and dental practices [ 142 ]. Moreover, several models have been reported to be inaccurate in predicting the clinical diagnosis. For example, an AI algorithm that can diagnose and classify chest X-rays using NLP to radiological records was developed [ 82 , 143 ]. These classifications were subsequently utilized in the training of a deep learning network to detect abnormalities in pictures, with a specific focus on recognizing a pneumothorax [ 144 ]. However, after a thorough examination, the presence of a chest tube in the majority of the reports identified as pneumothorax raised questions that the algorithm has been recognizing chest tubes instead of pneumothorax as envisioned [ 143 ]. Another example of non-interpretative results of a clinical AI-based system is DeepGestalt, a tool for analyzing facial dysmorphology. This tool performed poorly when identifying people with Down syndrome who were of African heritage (36.8%) compared to those who were of European origin (80%) [ 145 ]. The diagnoses of Down syndrome among people of African lineage increased to 94.7% when the model was retrained using cases of people with the condition [ 145 ]. Due to various marginalization in training datasets, genetic disease susceptibility modeling is also predisposed to differential performance across demographic groups [ 146 ]. Furthermore, it has been observed that while ML approaches may perform better in studies for developing disease risk prediction models, the presentation of the data may be more complex. There is also a possibility that the amount of computational time required by ML approaches varies depending on the size of the data [ 147 ]. Thus, it is crucial to acknowledge that the utilization of AI-based approaches will not always lead to improvised categorization or better prediction than present methods. AI is a tool that should be employed within the proper context to address a pertinent question or resolve a significant issue [ 148 ]. Similarly, in other biological fields such as agriculture, automation in practices employing AI and ML-based approaches leverages a lot of potential for sustainable farming. However, in the agricultural sector, the collection, analysis and utilization of data for productivity present a number of obstacles. Privacy and security of data are the two major challenges that farmers must address to survive in the digital age. In most cases, the farmers are uninformed of the collection, usage, and more concerningly, the purposes for which their personal details are being utilized [ 149 ]. Data mining allows corporations to rely on individuals in order to acquire massive agricultural data, which may be sufficient to develop and evaluate the behavioral and psychiatric pictures of the respondents [ 150 ]. To stop data from being misused, farmers require assurance that their information will be utilized to generate innovative ideas and agricultural solutions rather than to gain a competitive advantage. As mentioned elsewhere, the AI-based drone technology has emerged as a highly effective approach in agriculture [ 87 ]. However, drones, particularly those equipped with high-resolution lenses, infrared cameras, competent programs and sensors, are highly expensive for small farmers. Moreover, to operate drones, one needs authorization according to its operative and regulative provisions of the law of land [ 151 ]. Furthermore, weather imparts a huge influence on the operation of drones [ 152 ]. Traditional data mining methodologies are primarily developed for relational datasets; however, they are not completely adequate for geographically scattered data [ 153 ]. To revolutionize agriculture with AI-based technologies, innovative data mining approaches are needed.
In the industrial biotechnology sector, establishing defined and viable protocols for adopting an algorithm and assessing dataset size remain a major challenge. To design such protocols, it would be necessary to have a thorough knowledge of the effects/efficacy of various algorithms as well as training datasets to address numerous bioindustry challenges. Furthermore, increased accessibility, good documentation and superior data acquisition methods are still required to develop, operate and optimize bioenergy systems and bioreactor designs [ 128 ]. In some AI models, when the input is inadequate, particularly for large dimensional datasets, the algorithm may only recall every single variable as a special instance instead of learning the information, resulting in errors and lower training efficacy [ 154 ]. Additionally, numerous ANN-represented systems are frequently chastised for having black-box characteristics. Nonetheless, the paucity of comparative works across different AI–ML designs renders it challenging to present a clear direction for future studies or practical implementation [ 155 ]. There still exist challenges that need to be overcome including inefficient data integration which arises due to the diversity of the datasets inclusive of candidate data, metadata, processed data, raw data and lack of proper skill set and expertise related to the subject [ 156 ]. In this context, it is necessary to overcome these ambiguities by utilizing new AI algorithms to achieve a thorough alignment between the anticipated outcomes and the empirical studies [ 157 ]. Thus, more extensive datasets and relative studies are required to develop AI and ML-based models for real-time monitoring and control of bioreactors and bioprocesses.
6. Conclusions
One of the great achievements we have seen in the era of Industry 4.0 is the ability of a machine to replicate the capacities of living systems, particularly the intelligence of a human. The ability to recognize objects and make decisions is a crucial characteristic of biological systems. AI can currently recognize objects and make decisions using many of the cognitive and perceptual abilities of live systems. The potential of AI might be utilized to the biological world, including medical research, agriculture, and bio-based industries, for our sustainable way of life. The early prediction and identification of disease and its precise treatment based on personalized medicine even while the diseases are in asymptomatic conditions are examples of key areas in medical science that might benefit from AI. This would not only save millions of lives but also reduce medical costs. In addition to the medical field, AI-based efficient algorithms and programs have been recently developed to ensure effective inputs and outputs in farming, a practice known as precision farming. Agricultural practices such as soil management, water need analysis, exact modeling of fertilizer requirement, pesticides, insecticides, herbicides, yield projection and overall crop management could also be revolutionized by AI intervention. This would help to meet the world’s rising population’s demand for food. When we talk about large-scale production, many variable factors lead to increasing costs, which are major challenges. Recently, AI-based programs and computer models have proven to be very efficient at optimizing the suitable conditions to obtain the maximum desired product, whether for agricultural, medical, biotech, or lifestyle uses, at minimum cost. The efficient production of bio-enzymes is just one of such successes, and it is easy to envision how the biotech industry will be transformed by the application of AI, which will help to reduce production costs, one of the biggest challenges facing the industry today.
Acknowledgments
Authors are thankful to Amity University Jharkhand for the support provided under NTCC and PEARL Scheme.
Funding Statement
This research received no external funding.
Author Contributions
Conceptualization, D.K.P.; writing—original draft preparation, A.B.; writing—review and editing, A.B., S.K. and D.K.P.; supervision, D.K.P. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Informed consent statement, data availability statement, conflicts of interest.
The authors declare no conflict of interest.
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Secondary Menu
- Research Articles & Papers
Korunes, KL; Myers, RB; Hardy, R; Noor, MAF
Drosophila pseudoobscura is a classic model system for the study of evolutionary genetics and genomics. Given this long-standing interest, many genome sequences have accumulated for D. pseudoobscura and closely related species D. persimilis, D. miranda, and D. lowei. To facilitate the exploration… read more about this publication »
Zipple, MN; Roberts, EK; Alberts, SC; Beehner, JC
Bartoš et al. (2021; Mammal Review 51: 143–153; https://doi.org/10.1111/mam.12219) reviewed the mechanisms involved in the ‘Bruce effect’ – a phenomenon originally documented in inseminated female house mice Mus musculus, who block pregnancy following exposure to a novel (non-sire) male. They argue… read more about this publication »
Byrne, M; Koop, D; Strbenac, D; Cisternas, P; Yang, JYH; Davidson, PL; Wray, G
The molecular mechanisms underlying development of the pentameral body of adult echinoderms are poorly understood but are important to solve with respect to evolution of a unique body plan that contrasts with the bilateral body plan of other deuterostomes. As Nodal and BMP2/4 signalling is involved… read more about this publication »
Wang, Q; Xu, P; Sanchez, S; Duran, P; Andreazza, F; Isaacs, R; Dong, K
BackgroundInsects rely on their sense of smell to locate food and hosts, find mates and select sites for laying eggs. Use of volatile compounds, such as essential oils (EOs), to repel insect pests and disrupt their olfaction-driven behaviors has great practical significance in integrated pest… read more about this publication »
Castano-Duque, L; Ghosal, S; Quilloy, FA; Mitchell-Olds, T; Dixit, S
Rice production is shifting from transplanting seedlings to direct sowing of seeds. Following heavy rains, directly sown seeds may need to germinate under anaerobic environments, but most rice (Oryza sativa) genotypes cannot survive these conditions. To identify the genetic architecture of complex… read more about this publication »
Peng, L; Shan, X; Wang, Y; Martin, F; Vilgalys, R; Yuan, Z
Clitopilus hobsonii (Entolomataceae, Agaricales, Basidiomycetes) is a common soil saprotroph. There is also evidence that C. hobsonii can act as a root endophyte benefitting tree growth. Here, we report the genome assembly of C. hobsonii QYL-10, isolated from ectomycorrhizal root tips of Quercus… read more about this publication »
Yan, W; Wang, B; Chan, E; Mitchell-Olds, T
The genetic basis of flowering time changes across environments, and pleiotropy may limit adaptive evolution of populations in response to local conditions. However, little information is known about how genetic architecture changes among environments. We used genome-wide association studies (GWAS… read more about this publication »
Doak, DF; Waddle, E; Langendorf, RE; Louthan, AM; Isabelle Chardon, N; Dibner, RR; Keinath, DA; Lombardi, E; Steenbock, C; Shriver, RK; Linares, C; Begoña Garcia, M; Funk, WC; Fitzpatrick, SW; Morris, WF; Peterson, ML
Structured demographic models are among the most common and useful tools in population biology. However, the introduction of integral projection models (IPMs) has caused a profound shift in the way many demographic models are conceptualized. Some researchers have argued that IPMs, by explicitly… read more about this publication »
Kim, JH; Hilleary, R; Seroka, A; He, SY
A grand challenge facing plant scientists today is to find innovative solutions to increase global crop production in the context of an increasingly warming climate. A major roadblock to global food sufficiency is persistent loss of crops to plant diseases and insect infestations. The United… read more about this publication »
Yuan, M; Jiang, Z; Bi, G; Nomura, K; Liu, M; Wang, Y; Cai, B; Zhou, J-M; He, SY; Xin, X-F
The plant immune system is fundamental for plant survival in natural ecosystems and for productivity in crop fields. Substantial evidence supports the prevailing notion that plants possess a two-tiered innate immune system, called pattern-triggered immunity (PTI) and effector-triggered immunity (… read more about this publication »
Benito-Kwiecinski, S; Giandomenico, SL; Sutcliffe, M; Riis, ES; Freire-Pritchett, P; Kelava, I; Wunderlich, S; Martin, U; Wray, GA; McDole, K; Lancaster, MA
The human brain has undergone rapid expansion since humans diverged from other great apes, but the mechanism of this human-specific enlargement is still unknown. Here, we use cerebral organoids derived from human, gorilla, and chimpanzee cells to study developmental mechanisms driving evolutionary… read more about this publication »
Lofgren, LA; Nguyen, NH; Vilgalys, R; Ruytinx, J; Liao, H-L; Branco, S; Kuo, A; LaButti, K; Lipzen, A; Andreopoulos, W; Pangilinan, J; Riley, R; Hundley, H; Na, H; Barry, K; Grigoriev, IV; Stajich, JE; Kennedy, PG
While there has been significant progress characterizing the 'symbiotic toolkit' of ectomycorrhizal (ECM) fungi, how host specificity may be encoded into ECM fungal genomes remains poorly understood. We conducted a comparative genomic analysis of ECM fungal host specialists and generalists,… read more about this publication »
Mitchell, RM; Ames, GM; Wright, JP
Background and aimsUnderstanding impacts of altered disturbance regimes on community structure and function is a key goal for community ecology. Functional traits link species composition to ecosystem functioning. Changes in the distribution of functional traits at community scales in response to… read more about this publication »
DeMarche, ML; Bailes, G; Hendricks, LB; Pfeifer-Meister, L; Reed, PB; Bridgham, SD; Johnson, BR; Shriver, R; Waddle, E; Wroton, H; Doak, DF; Roy, BA; Morris, WF
Spatial gradients in population growth, such as across latitudinal or elevational gradients, are often assumed to primarily be driven by variation in climate, and are frequently used to infer species' responses to climate change. Here, we use a novel demographic, mixed-model approach to dissect the… read more about this publication »
Shaw, EC; Fowler, R; Ohadi, S; Bayly, MJ; Barrett, RA; Tibbits, J; Strand, A; Willis, CG; Donohue, K; Robeck, P; Cousens, RD
Aim: If we are able to determine the geographic origin of an invasion, as well as its known area of introduction, we can better appreciate the innate environmental tolerance of a species and the strength of selection for adaptation that colonizing populations have undergone. It also enables us to… read more about this publication »
Rushworth, CA; Mitchell-Olds, T
Despite decades of research, the evolution of sex remains an enigma in evolutionary biology. Typically, research addresses the costs of sex and asexuality to characterize the circumstances favoring one reproductive mode. Surprisingly few studies address the influence of common traits that are, in… read more about this publication »
Jorge, JF; Bergbreiter, S; Patek, SN
Small organisms can produce powerful, sub-millisecond impacts by moving tiny structures at high accelerations. We developed and validated a pendulum device to measure the impact energetics of microgram-sized trap-jaw ant mandibles accelerated against targets at 105 m s-2 Trap-jaw ants (… read more about this publication »
Markunas, AM; Manivannan, PKR; Ezekian, JE; Agarwal, A; Eisner, W; Alsina, K; Allen, HD; Wray, GA; Kim, JJ; Wehrens, XHT; Landstrom, AP
Long QT syndrome (LQTS) is a genetic disease resulting in a prolonged QT interval on a resting electrocardiogram, predisposing affected individuals to polymorphic ventricular tachycardia and sudden death. Although a number of genes have been implicated in this disease, nearly one in four… read more about this publication »
Stone, DF; Mccune, B; Pardo-De La Hoz, CJ; Magain, N; Miadlikowska, J
The new genus Sinuicella, an early successional lichen, was found on bare soil in Oregon, USA. The thallus is minute fruticose, grey to nearly black, branching isotomic dichotomous, branches round, 20-90 μm wide in water mount. The cortex is composed of interlocking cells shaped like jigsaw puzzle… read more about this publication »
Oita, S; Ibáñez, A; Lutzoni, F; Miadlikowska, J; Geml, J; Lewis, LA; Hom, EFY; Carbone, I; U'Ren, JM; Arnold, AE
Understanding how species-rich communities persist is a foundational question in ecology. In tropical forests, tree diversity is structured by edaphic factors, climate, and biotic interactions, with seasonality playing an essential role at landscape scales: wetter and less seasonal forests… read more about this publication »
Hibshman, JD; Webster, AK; Baugh, LR
Standard laboratory culture of Caenorhabditis elegans utilizes solid growth media with a bacterial food source. However, this culture method limits control of food availability and worm population density, factors that impact many life-history traits. Here, we describe liquid-culture protocols for… read more about this publication »
Caves, EM; Green, PA; Zipple, MN; Bharath, D; Peters, S; Johnsen, S; Nowicki, S
AbstractSensory systems are predicted to be adapted to the perception of important stimuli, such as signals used in communication. Prior work has shown that female zebra finches perceive the carotenoid-based orange-red coloration of male beaks-a mate choice signal-categorically. Specifically,… read more about this publication »
Reed, PB; Peterson, ML; Pfeifer-Meister, LE; Morris, WF; Doak, DF; Roy, BA; Johnson, BR; Bailes, GT; Nelson, AA; Bridgham, SD
Predicting species' range shifts under future climate is a central goal of conservation ecology. Studying populations within and beyond multiple species' current ranges can help identify whether demographic responses to climate change exhibit directionality, indicative of range shifts, and whether… read more about this publication »
Sallee, JL; Crawford, JM; Singh, V; Kiehart, DP
Actin filament crosslinking, bundling and molecular motor proteins are necessary for the assembly of epithelial projections such as microvilli, stereocilia, hairs, and bristles. Mutations in such proteins cause defects in the shape, structure, and function of these actin - based protrusions. One… read more about this publication »
- Duke Biology’s Mission Statement
- AJED Annual and Semester Reports
- AJED Meeting Notes
- Biology Cultural Association (BCA)
- Inclusion, Diversity, Equity, and Antiracism Committee (IDEA)
- Learning from Baboons: Dr. Susan Alberts
- Extremophiles and Systems Biology: Dr. Amy Schmid
- How Cells Manage Stress: Dr. Gustavo Silva
- Predator-Prey Interactions in a Changing World: Dr. Jean Philippe Gibert
- Exploring the Extracellular Matrix: Dr. David Sherwood
- Cell Division's Missing Link: Dr. Masayuki Onishi
- Listening in to Birdsong: Dr. Steve Nowicki
- Biogeochemistry as Ecosystem Accounting: Dr. Emily Bernhardt
- Building a Dynamic Nervous System: Dr. Pelin Volkan
- Investigating a Key Plant Hormone: Dr. Lucia Strader
- Imagining Visual Ecology: Dr. Sönke Johnsen
- Outreach Opportunities Across the Triangle
- Job Opportunities
- Location & Contact
- Frequently Asked Questions
- Learning Outcomes
- Major Requirements
- Anatomy, Physiology & Biomechanics
- Animal Behavior
- Biochemistry
- Cell & Molecular Biology
- Evolutionary Biology
- Marine Biology
- Neurobiology
- Pharmacology
- Plant Biology
- Minor Requirements
- Biology IDM
- List of Biology Advisors
- Guide for First-Year Students
- Transfer Credit
- Application & Deadlines
- Supervisor & Faculty Reader
- Thesis Guidelines
- Honors Poster
- Past Student Projects
- Study Away Opportunities
- Finding a Research Mentor
- Project Guidelines
- Getting Registered
- Writing Intensive Study
- Independent Study Abroad
- Summer Opportunities
- Departmental Awards
- Biology Majors Union
- Commencement 2024
- Trinity Ambassadors
- Degree Programs
- Ph.D. Requirements
- How to Apply
- Financial Aid
- Living in Durham
- Where Our Students Go
- Milestones Toward Ph.D.
- Graduate School Fellowships
- Useful Resources
- Concurrent Biology Master of Science
- En Route Biology Masters of Science
- Form Library
- Mentorship Expectations
- On Campus Resources
- Fellowships & Jobs
- Meet Our Postdocs
- Department Research Areas
- Research Facilities
- Duke Postdoctoral Association
- All Courses
- Biological Structure & Function Courses
- Ecology Courses
- Organismal Diversity Courses
- Alternate Elective Courses
- Primary Faculty
- Secondary Faculty
- Graduate Faculty
- Emeritus Faculty
- Graduate Students
- Department Staff
- Faculty Research Labs
- Developmental Biology
- Ecology & Population Biology
- Neuroscience
- Organismal Biology & Behavior
- Systematics
- Botany Plot
- Field Station
- Pest Management Protocols
- Research Greenhouses
- Centers/Research Groups
- Biology Writes
- Alumni Profiles
- For Current Students
- Assisting Duke Students
Academia.edu no longer supports Internet Explorer.
To browse Academia.edu and the wider internet faster and more securely, please take a few seconds to upgrade your browser .
- We're Hiring!
- Help Center
- Most Cited Papers
- Most Downloaded Papers
- Newest Papers
- Save to Library
- Last »
- Ecology Follow Following
- Evolutionary Biology Follow Following
- Conservation Biology Follow Following
- Biodiversity Follow Following
- Evolution Follow Following
- Zoology Follow Following
- Behavioral Ecology Follow Following
- Systematics (Taxonomy) Follow Following
- Community Ecology Follow Following
- Phylogenetics Follow Following
Enter the email address you signed up with and we'll email you a reset link.
- Academia.edu Publishing
- We're Hiring!
- Help Center
- Find new research papers in:
- Health Sciences
- Earth Sciences
- Cognitive Science
- Mathematics
- Computer Science
- Academia ©2024
- How it works
Published by Robert Bruce at August 29th, 2023 , Revised On September 5, 2023
Biology Research Topics
Are you in need of captivating and achievable research topics within the field of biology? Your quest for the best biology topics ends right here as this article furnishes you with 100 distinctive and original concepts for biology research, laying the groundwork for your research endeavor.
Table of Contents
Our proficient researchers have thoughtfully curated these biology research themes, considering the substantial body of literature accessible and the prevailing gaps in research.
Should none of these topics elicit enthusiasm, our specialists are equally capable of proposing tailor-made research ideas in biology, finely tuned to cater to your requirements.
Thus, without further delay, we present our compilation of biology research topics crafted to accommodate students and researchers.
Research Topics in Marine Biology
- Impact of climate change on coral reef ecosystems.
- Biodiversity and adaptation of deep-sea organisms.
- Effects of pollution on marine life and ecosystems.
- Role of marine protected areas in conserving biodiversity.
- Microplastics in marine environments: sources, impacts, and mitigation.
Biological Anthropology Research Topics
- Evolutionary implications of early human migration patterns.
- Genetic and environmental factors influencing human height variation.
- Cultural evolution and its impact on human societies.
- Paleoanthropological insights into human dietary adaptations.
- Genetic diversity and population history of indigenous communities.
Biological Psychology Research Topics
- Neurobiological basis of addiction and its treatment.
- Impact of stress on brain structure and function.
- Genetic and environmental influences on mental health disorders.
- Neural mechanisms underlying emotions and emotional regulation.
- Role of the gut-brain axis in psychological well-being.
Cancer Biology Research Topics
- Targeted therapies in precision cancer medicine.
- Tumor microenvironment and its influence on cancer progression.
- Epigenetic modifications in cancer development and therapy.
- Immune checkpoint inhibitors and their role in cancer immunotherapy.
- Early detection and diagnosis strategies for various types of cancer.
Also read: Cancer research topics

Cell Biology Research Topics
- Mechanisms of autophagy and its implications in health and disease.
- Intracellular transport and organelle dynamics in cell function.
- Role of cell signaling pathways in cellular response to external stimuli.
- Cell cycle regulation and its relevance to cancer development.
- Cellular mechanisms of apoptosis and programmed cell death.
Developmental Biology Research Topics
- Genetic and molecular basis of limb development in vertebrates.
- Evolution of embryonic development and its impact on morphological diversity.
- Stem cell therapy and regenerative medicine approaches.
- Mechanisms of organogenesis and tissue regeneration in animals.
- Role of non-coding RNAs in developmental processes.
Also read: Education research topics
Human Biology Research Topics
- Genetic factors influencing susceptibility to infectious diseases.
- Human microbiome and its impact on health and disease.
- Genetic basis of rare and common human diseases.
- Genetic and environmental factors contributing to aging.
- Impact of lifestyle and diet on human health and longevity.
Molecular Biology Research Topics
- CRISPR-Cas gene editing technology and its applications.
- Non-coding RNAs as regulators of gene expression.
- Role of epigenetics in gene regulation and disease.
- Mechanisms of DNA repair and genome stability.
- Molecular basis of cellular metabolism and energy production.
Research Topics in Biology for Undergraduates
- 41. Investigating the effects of pollutants on local plant species.
- Microbial diversity and ecosystem functioning in a specific habitat.
- Understanding the genetics of antibiotic resistance in bacteria.
- Impact of urbanization on bird populations and biodiversity.
- Investigating the role of pheromones in insect communication.
Synthetic Biology Research Topics
- Design and construction of synthetic biological circuits.
- Synthetic biology applications in biofuel production.
- Ethical considerations in synthetic biology research and applications.
- Synthetic biology approaches to engineering novel enzymes.
- Creating synthetic organisms with modified functions and capabilities.
Animal Biology Research Topics
- Evolution of mating behaviors in animal species.
- Genetic basis of color variation in butterfly wings.
- Impact of habitat fragmentation on amphibian populations.
- Behavior and communication in social insect colonies.
- Adaptations of marine mammals to aquatic environments.
Also read: Nursing research topics
Best Biology Research Topics
- Unraveling the mysteries of circadian rhythms in organisms.
- Investigating the ecological significance of cryptic coloration.
- Evolution of venomous animals and their prey.
- The role of endosymbiosis in the evolution of eukaryotic cells.
- Exploring the potential of extremophiles in biotechnology.
Biological Psychology Research Paper Topics
- Neurobiological mechanisms underlying memory formation.
- Impact of sleep disorders on cognitive function and mental health.
- Biological basis of personality traits and behavior.
- Neural correlates of emotions and emotional disorders.
- Role of neuroplasticity in brain recovery after injury.
Biological Science Research Topics:
- Role of gut microbiota in immune system development.
- Molecular mechanisms of gene regulation during development.
- Impact of climate change on insect population dynamics.
- Genetic basis of neurodegenerative diseases like Alzheimer’s.
- Evolutionary relationships among vertebrate species based on DNA analysis.
Biology Education Research Topics
- Effectiveness of inquiry-based learning in biology classrooms.
- Assessing the impact of virtual labs on student understanding of biology concepts.
- Gender disparities in science education and strategies for closing the gap.
- Role of outdoor education in enhancing students’ ecological awareness.
- Integrating technology in biology education: challenges and opportunities.
Biology-Related Research Topics
- The intersection of ecology and economics in conservation planning.
- Molecular basis of antibiotic resistance in pathogenic bacteria.
- Implications of genetic modification of crops for food security.
- Evolutionary perspectives on cooperation and altruism in animal behavior.
- Environmental impacts of genetically modified organisms (GMOs).
Biology Research Proposal Topics
- Investigating the role of microRNAs in cancer progression.
- Exploring the effects of pollution on aquatic biodiversity.
- Developing a gene therapy approach for a genetic disorder.
- Assessing the potential of natural compounds as anti-inflammatory agents.
- Studying the molecular basis of cellular senescence and aging.
Biology Research Topic Ideas
- Role of pheromones in insect mate selection and behavior.
- Investigating the molecular basis of neurodevelopmental disorders.
- Impact of climate change on plant-pollinator interactions.
- Genetic diversity and conservation of endangered species.
- Evolutionary patterns in mimicry and camouflage in organisms.
Biology Research Topics for Undergraduates
- Effects of different fertilizers on plant growth and soil health.
- Investigating the biodiversity of a local freshwater ecosystem.
- Evolutionary origins of a specific animal adaptation.
- Genetic diversity and disease susceptibility in human populations.
- Role of specific genes in regulating the immune response.
Cell and Molecular Biology Research Topics
- Molecular mechanisms of DNA replication and repair.
- Role of microRNAs in post-transcriptional gene regulation.
- Investigating the cell cycle and its control mechanisms.
- Molecular basis of mitochondrial diseases and therapies.
- Cellular responses to oxidative stress and their implications in ageing.
These topics cover a broad range of subjects within biology, offering plenty of options for research projects. Remember that you can further refine these topics based on your specific interests and research goals.
Frequently Asked Questions
What are some good research topics in biology?
A good research topic in biology will address a specific problem in any of the several areas of biology, such as marine biology, molecular biology, cellular biology, animal biology, or cancer biology.
A topic that enables you to investigate a problem in any area of biology will help you make a meaningful contribution.
How to choose a research topic in biology?
Choosing a research topic in biology is simple.
Follow the steps:
- Generate potential topics.
- Consider your areas of knowledge and personal passions.
- Conduct a thorough review of existing literature.
- Evaluate the practicality and viability.
- Narrow down and refine your research query.
- Remain receptive to new ideas and suggestions.
Who Are We?
For several years, Research Prospect has been offering students around the globe complimentary research topic suggestions. We aim to assist students in choosing a research topic that is both suitable and feasible for their project, leading to the attainment of their desired grades. Explore how our services, including research proposal writing , dissertation outline creation, and comprehensive thesis writing , can contribute to your college’s success.
You May Also Like
Welcome to the most comprehensive resource page of climate change research topics, a crucial field of study central to understanding […]
How many universities in Canada? Over 100 private universities and 96 public ones, the top ones include U of T, UBC, and Western University.
A preliminary literature review is an initial exploration of existing research on a topic, setting the foundation for in-depth study.
Ready to place an order?
USEFUL LINKS
Learning resources, company details.
- How It Works
Automated page speed optimizations for fast site performance
Interview with Gerd B. Müller on Theoretical Biology
- Perspective
- Published: 09 October 2023
- Volume 16 , pages 381–394, ( 2023 )
Cite this article
- Kalevi Kull 1
159 Accesses
Explore all metrics
The topics discussed in the interview cover the development and activities of the Konrad Lorenz Institute for Evolution and Cognition Research as one of the most important theoretical biology centers in the world, the reasons for its inspiring atmosphere, as well as the development of the interests and research work of its longtime president Gerd B. Müller. An important part of this is the work on a revised theoretical framework of evolution, the Extended Evolutionary Synthesis. We also talk about the place of biosemiotics in biology and about the situation and future perspectives of theoretical biology in general. With this interview we celebrate Gerd Müller’s 70th birthday.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions

Similar content being viewed by others

Philosophy of Evolutionary Biology
A. P. Rasnitsyn
In Memoriam Werner Callebaut
Giovanni Boniolo

Evolutionary Biology and Ethics
See also his autobiographical sketch: Müller, 2011 .
About this important conference, see a review in Kull, 2016 .
See, for instance, an overview in Kull & Favareau, 2022 . In the context of the extended evolutionary synthesis, see also Kull, 2022 .
See also Laubichler & Müller, 2007 .
See Müller, 2007 .
EES. See Pigliucci & Müller, 2010 .
See Müller, 1985 .
See Müller, 2021 .
See Müller, 2003 .
See, for instance, Newman et al., 2006 .
Streicher et al., 2000 .
Lange et al., 2018 .
Müller et al., 1996 .
Nuño de la Rosa & Müller, 2021 .
Müller & Abouheif, 2021 .
Müller, 2017a .
About this club, see Peterson, 2016 .
Bertalanffy, 1933 .
Müller, 2017b .
Extended Evolutionary Synthesis. For a brief summary see Müller, 2017c .
Riedl, 1978 .
Müggenburg, 2018 .
Cf. Goy & Watkins, 2014 .
The volumes are not numbered.
Quote: "Today theoretical biology has genetic, developmental, and evolutionary components, the central connective themes in modern biology, but also includes relevant aspects of computational biology, semiotics, and cognition research, and extends to the naturalistic philosophy of sciences" (Müller et al., 2003 : vii).
Or: "No new truth will declare itself from inside a heap of facts" (Medawar 1979 : 71).
Bertalanffy, L. von (1933). Modern theories of development: An introduction to theoretical biology (trans and adapted by J. H. Woodger). Oxford University Press.
Goy, I., & Watkins, E. (2014). Kant’s theory of biology . De Gruyter.
Book Google Scholar
Kull, K. (2016). What kind of evolutionary biology suits cultural research. Sign Systems Studies, 44 (4), 634–647.
Article Google Scholar
Kull, K. (2022). The aim of extended synthesis is to include semiosis. Theoretical Biology Forum, 115 (1/2), 119–132.
PubMed Google Scholar
Kull, K. & Favareau, D. (2022). Semiotics in general biology. In: Pelkey, J. & Walsh Matthews, S. (Eds.), Bloomsbury semiotics , vol. 2: Semiotics in the natural and technical sciences (pp. 35–56). Bloomsbury Academic.
Lange, A., Nemeschkal, H. L., & Müller, G. B. (2018). A threshold model for polydactyly. Progress in Biophysics and Molecular Biology, 137 , 1–11.
Article PubMed Google Scholar
Laubichler, M. D., & Müller, G. B. (2007). Models in theoretical biology. In: Laubichler, M. D., & Müller, G. B. (Eds.), Modeling biology: Structures, behaviors, evolution (pp. 3–10). (The Vienna Series in Theoretical Biology.). The MIT Press.
Medawar, P. B. (1979). Advice to a young scientist . Harper & Row.
Google Scholar
Müggenburg, J. (2018). Lebhafte Artefakte: Heinz von Foerster und die Maschinen des Biological Computer Laboratory . Konstanz University Press.
Müller, G. B. (1985). Experimentelle Untersuchungen zur Theorie des epigenetischen Systems. In J. A. Ott, G. P. Wagner, & F. M. Wuketits (Eds.), Evolution, Ordnung und Erkenntnis (pp. 92–96). Paul Parey.
Müller, G. B. (2003). Homology: The evolution of morphological organization. In: Müller, G. B, Newman, S. A. (Eds.), Origination of organismal form: Beyond the gene in the developmental and evolutionary biology (pp. 51–69). (The Vienna Series in Theoretical Biology.). The MIT Press.
Müller, G. B. (2007). Evo-devo: Extending the evolutionary synthesis. Nature Reviews Genetics, 8 (12), 943–949.
Müller, G. B. (2011). Bio . Evolution & Development, 13 (3), 243–246.
Müller, G. B. (2017a). The substance of form: Hans Przibram’s quest for biological experiment, quantification, and theory. In: Müller, Gerd B. (Ed.), Vivarium – experimental, quantitative, and theoretical biology at Vienna’s Biologische Versuchsanstalt (pp. 135–164) . (The Vienna Series in Theoretical Biology.). The MIT Press.
Müller, G. B. (Ed.) (2017b). Vivarium: Experimental, quantitative, and theoretical biology at Vienna’s Biologische Versuchsanstalt. (The Vienna Series in Theoretical Biology.). The MIT Press.
Müller, G. B. (2017c). Why an extended evolutionary synthesis is necessary. Interface Focus, 7 (5), 1–11.
Müller, G. B. (2021). Developmental innovation and phenotypic novelty. In L. Nuño de la Rosa & G. B. Müller (Eds.), Evolutionary developmental biology: A reference guide (Vol. 2, pp. 69–84). Springer.
Chapter Google Scholar
Müller, G. B., & Abouheif, E. (2021). Evolutionary developmental biology . Oxford Bibliographies in Evolutionary Biology.
Müller, G. B., Streicher, J., & Müller, R. J. (1996). Homeotic duplication of the pelvic body segment in regenerating tadpole tails induced by retinoic acid. Development, Genes and Evolution, 206 (5), 344–348.
Müller, G. B., Wagner, G. P., & Callebaut, W. (2003). Series foreword. In: L. Tommasi, M. A. Peterson, & L. Nadel (Eds.), Cognitive biology: Evolutionary and developmental perspectives on mind, brain and behavior (pp. vii–viii). (The Vienna Series in Theoretical Biology.). The MIT Press.
Newman, S. A., Forgacs, G., & Müller, G. B. (2006). Before programs: The physical origination of multicellular forms. The International Journal of Developmental Biology, 50 (2/3), 289–299.
Article CAS Google Scholar
Nuño de la Rosa, L., & Müller, G. B. (Eds.). (2021). Evolutionary developmental biology: A reference guide . Springer.
Peterson, E. L. (2016). The life organic: The theoretical biology club and the roots of epigenetics . University of Pittsburgh Press.
Pigliucci, M., & Müller, G. B. (Eds.). (2010). Evolution: The extended synthesis . The MIT Press.
Riedl, R. (1978). Order in living organisms: A systems analysis of evolution . (trans by Jefferies, R. P. S.). Wiley.
Streicher, J., Donat, M. A., Strauss, B., Spörle, R., Schughart, K., & Müller, G. B. (2000). Computer-based three-dimensional visualization of developmental gene expression. Nature Genetics, 25 (2), 147–152.
Article CAS PubMed Google Scholar
Download references
Acknowledgements
Many thanks to Gerd Müller for his excellent work in theoretical biology and congratulations on his 70th birthday.
Author information
Authors and affiliations.
Department of Semiotics, University of Tartu, Tartu, Estonia
Kalevi Kull
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Kalevi Kull .
Ethics declarations
Conflict of interests.
The author declares no conflict of interests.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Kull, K. Interview with Gerd B. Müller on Theoretical Biology. Biosemiotics 16 , 381–394 (2023). https://doi.org/10.1007/s12304-023-09543-w
Download citation
Received : 31 August 2023
Accepted : 11 September 2023
Published : 09 October 2023
Issue Date : December 2023
DOI : https://doi.org/10.1007/s12304-023-09543-w
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Austrian biology
- Developmental biology
- Extended evolutionary synthesis
- History of biology
- Konrad Lorenz Institute
- Theoretical biology
- Find a journal
- Publish with us
- Track your research

- Dean’s Message
- Faculty Members
- Administrative Staff
- Lab Managers
- Useful links for Faculty
- Dear Alumni
- Faculty Positions
- Testimonials
- Why Biology – Information for Applicants
- Undergraduate programs
- Curriculum catalog
- Student Dormitories
- Request form for Grade Reevaluation
- “MILUIM” date and special date
- List of Advisors
- Graduates tell
- Graduate Study Programs
- Search for a Supervisor
- Scholarships and Housing
- Grad Courses
- TA Positions for Grad Students
- TA Positions for Foreign Students
- Academic Requirements for a Ph.D. Degree in Biology
- Academic Requirements for an M.Sc. Degree in Biology
- Scholarships
- Research Opportunities
- Research Areas
- Research infrastructures
- faculty members by research areas
- Postdoctoral
- About the program
- How to Apply
- Contact Details

The winner of February 2024 article is Guy Levin from Prof. Gadi Schuster’s lab. The article was published in New Phytologist.

SPOTLIGHT ON RESEARCH PAPER OF THE MONTH FEB 2024
The Committee for Graduate Studies in the Faculty of Biology selects the leading scientific article each month from all the scientific articles published for that month. We are pleased to announce that the winner of February 2024 article is Guy Levin from Prof. Gadi Schuster’s lab. The article was published in New Phytologist. On the occasion of the win, we asked Guy Levin to provide us with some interesting details about the study, the path that led to the research, and a bit about him.
Hi, could you introduce yourself in a few words? Hi, I’m Guy, 34, Married to Nofar and the father of a beautiful baby-girl, Ella. Could you explain what Prof. Schuster’s Laboratory is all about? Our lab does research in various fields. Starting from studying nucleases (RNAse J), through photosynthesis in extreme conditions and up to producing electricity via photosynthesis.
Could you tell us about your current article/research what was the main purpose of the research and what did you discover? My research focuses on the question of how Chlorella ohadii , a green microalga, can thrive in the desert under light intensities that are fatal to most organisms. This work is a continuation of my previous manuscript, in which we found that a main photosynthetic complex, photosystem I, undergoes phosphorylation. Phosphorylation of photosynthetic proteins has many roles, however, the role of Photosystem I phosphorylation is unknown, as well as the identity of the specific subunits that are phosphorylated. In this manuscript, we successfully mapped all of the phosphorylation sites of photosystem I, in unprecedented detail. Additionally, we analyzed the effects of light intensity on the identity of the phosphorylated proteins, to detect phosphorylation sites which are important for acclimation to high-light growing conditions. Moreover, we used molecular dynamics to analyze the possible roles of some of these phosphorylation sites and we demonstrated why they may be important for the stability and function of the photosynthetic proteins.
Can you elaborate on the importance of the discovery? How will it serve you and what directions does it take? What is the application of the discovery (domains, solutions)? This work is another important piece in the puzzle we are putting together during my PhD research. The ultimate goal of my research is to understand the molecular mechanisms that enable Chlorella ohadii to thrive under extremely high light intensities. Then, I aim to use this knowledge and introduce the light-tolerant phenotype to important crops like rice, via genetic tools. Doing so will enable us to expand the niche of the crops and enhance their growth and yield.
N ame 3 prominent tools that you received in the laboratory during your work and studies.
- Time management
- I have learned to keep an open mind and be ready for changes. This is important because in science things often don’t go our way.
- Never give up. And this is probably the best advice I can give to young students.
When you are not “doing” science, what do you do? My baby was just born a month ago, so I spend most of my time with my family. In addition, I’m a massive Liverpool fan and also love to scuba-dive and travel across our beautiful country.
what are your plans for the future of your career? Soon I will be completing my Ph.D. research and will start postdoc research in the US. Hopefully, after that, I can open my lab here in the faculty.
➡ A link to the full article : https://doi.org/10.1111/nph.19603 ➡ A link to the Prof. Schuster’s lab site: https://schuster.net.technion.ac.il ➡ To Prof. Gadi Schuster’s page: https://biology.technion.ac.il/member/%d7%a9%d7%95%d7%a1%d7%98%d7%a8
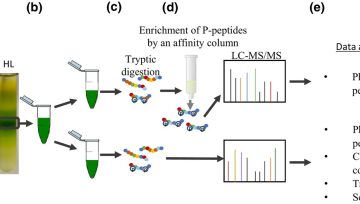
All News and Events

- Declaration of accessibility
- Biology Major
- Molecular Biochemistry
- Research-Oriented Honors Program
- Biology and Materials Engineering
- Biochemical Engineering
- Computer Science with Concentration in Bioinformatics
- M.Sc. Biology
- Transfer to the Direct Ph.D. Program
- Honors Rotation Program for Excellent Students in the Combined Ph.D. Program
- Doctoral (Ph.D.) Studies
- Life Sciences MBA as a Minor Combined with Ph.D. Studies

Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 14 March 2024
Edible mycelium bioengineered for enhanced nutritional value and sensory appeal using a modular synthetic biology toolkit
- Vayu Maini Rekdal 1 , 2 , 3 ,
- Casper R. B. van der Luijt ORCID: orcid.org/0000-0001-5978-7731 3 , 4 , 5 , 6 ,
- Yan Chen 3 , 6 ,
- Ramu Kakumanu 3 , 6 ,
- Edward E. K. Baidoo 3 , 6 ,
- Christopher J. Petzold ORCID: orcid.org/0000-0002-8270-5228 3 , 6 ,
- Pablo Cruz-Morales 4 &
- Jay D. Keasling ORCID: orcid.org/0000-0003-4170-6088 1 , 3 , 4 , 6 , 7 , 8
Nature Communications volume 15 , Article number: 2099 ( 2024 ) Cite this article
10k Accesses
297 Altmetric
Metrics details
- Food microbiology
- Genetic engineering
- Metabolic engineering
- Synthetic biology
Filamentous fungi are critical in the transition to a more sustainable food system. While genetic modification of these organisms has promise for enhancing the nutritional value, sensory appeal, and scalability of fungal foods, genetic tools and demonstrated use cases for bioengineered food production by edible strains are lacking. Here, we develop a modular synthetic biology toolkit for Aspergillus oryzae , an edible fungus used in fermented foods, protein production, and meat alternatives. Our toolkit includes a CRISPR-Cas9 method for gene integration, neutral loci, and tunable promoters. We use these tools to elevate intracellular levels of the nutraceutical ergothioneine and the flavor-and color molecule heme in the edible biomass. The strain overproducing heme is red in color and is readily formulated into imitation meat patties with minimal processing. These findings highlight the promise of synthetic biology to enhance fungal foods and provide useful genetic tools for applications in food production and beyond.
Similar content being viewed by others
The microbial food revolution
Alicia E. Graham & Rodrigo Ledesma-Amaro
Synthetic biology strategies for microbial biosynthesis of plant natural products
Aaron Cravens, James Payne & Christina D. Smolke
New colours for old in the blue-cheese fungus Penicillium roqueforti
Matthew M. Cleere, Michaela Novodvorska, … Paul S. Dyer
Introduction
The global food system has been identified as one of the major contributors to climate change. Food production is responsible for an estimated one-third of global greenhouse gas emissions and contributes to widespread environmental degradation, biodiversity loss, and the emergence of new diseases 1 , 2 , 3 . Transitioning food production away from resource-intensive industrial animal agriculture toward alternative methods, including microbial processes, is critical for mitigating these negative planetary impacts and sustainably feeding a growing global population that is estimated to reach over 9 billion by 2050 1 , 2 , 4 , 5 , 6 , 7 . Among other applications, microbes can be used for upcycling byproducts 8 , as hosts for production of environmentally taxing small molecules and proteins 9 , 10 , and for producing nutritious biomass that can be consumed directly 11 . Compared to animal agriculture, microbial food production can offer increased resource efficiency and safety, more precise control of production, reduced animal suffering, and a reduced environmental footprint 5 .
Filamentous fungi, a diverse group of microorganisms that includes molds and mushrooms, have several advantages compared to other hosts for microbially based food production 12 (Fig. 1A ). In addition to the historical use of many fungi for safe and delicious fermented foods 13 , the naturally high secretory capacity of these organisms makes them powerful hosts for production of proteins for food and other uses 14 . Additionally, many fungi rapidly degrade and grow on complex substrates such as food byproducts or lignocellulose, which can alleviate the cost and environmental burden associated with highly purified substrates such as glucose 9 , 15 . Finally, owing to its filamentous morphology which mimics the structure of animal muscle, fungal biomass (mycelium) can be formulated into meat alternatives with convincing textures (mycoprotein), and used as scaffolds for adherent animal cells in cellular agriculture 11 . A recent Life Cycle Assessment revealed that substituting 20% of animal protein with mycoprotein by 2050 could lower methane emissions as well as reduce deforestation and associated CO 2 emissions by half, underscoring the concrete environmental benefits of fungal foods 6 .
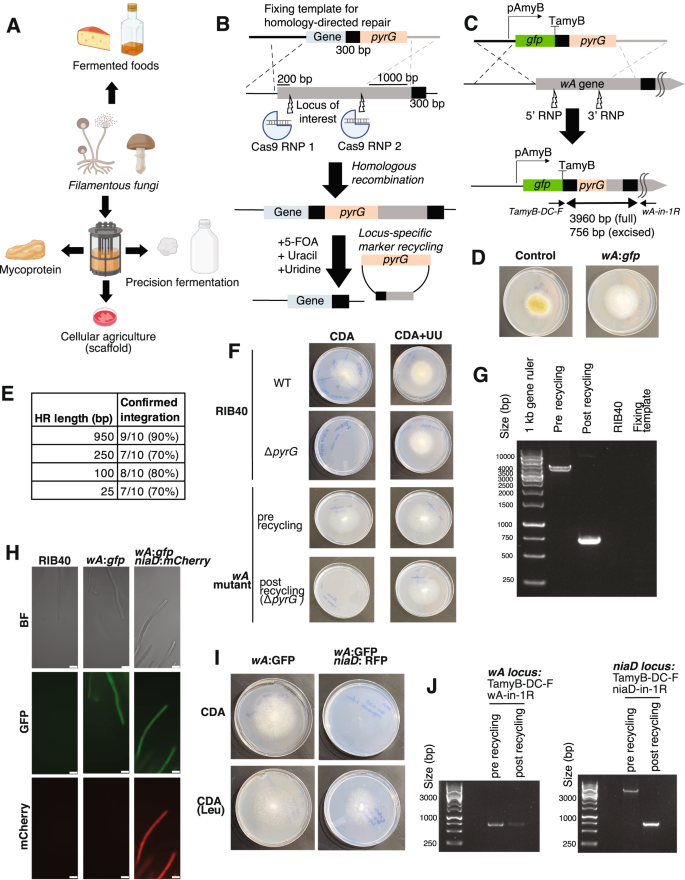
A Fungal applications in sustainable food production. The figure was created was created using BioRender ( http://BioRender.com ). B Strategy for RNP-based CRISPR-Cas9 editing. Upon integration at the correct locus, the pyrG selection marker becomes flanked by two identical 300-bp sequences. Counter-selection using 5-fluoroorotic acid (5-FOA) allows locus-specific marker excision. C Strategy for integration of a GFP-expression cassette (pAmyB promoter) at the wA locus, which controls spore pigmentation, in A. oryzae RIB40. The two squiggly lines indicate that not the whole 6.7-kb wA gene is shown. Primers TamyB-DC-F (on fixing template) and wA-in-1R (on chromosome) were used to confirm successful insertion. Ladder is Generuler 1 kb ladder (Thermo Scientific). D The wA : gfp transformant has white conidia instead of the yellow-green conidial pigmentation of the RIB40 strain (grown on PDA medium). E Efficiency of gene integration at the wA locus as a function of the homology arm length in the fixing template. 10 colonies were randomly selected at each length and subjected to colony PCR. F Following pyrG marker recycling from the wA locus the strain displays uracil/uridine auxotrophy when grown on CDA medium. UU = uracil and uridine supplementation G The auxotrophy was accompanied by clear marker loop out from the wA locus. Ladder is Generuler 1 kb ladder (Thermo Scientific). H The looped-out wA:gfp strain (Δ pyrG) was transformed with an mCherry expression cassette targeted at the niaD locus, which controls nitrate assimilation. Microscopy in the wA:gfp and niaD:mCherry strain confirmed the expected pattern of protein expression. Scale bar = 25 µm. I The resulting transformant only grows with leucine (CDA-Leu) as the nitrogen source instead of nitrate (CDA), as indicated by the colony radiating from the center of the plate. J The wA:gfp and niaD:mCherry strain was subjected to marker recycling from the niaD locus. PCR confirmation indicates that marker recycling was successful from the niaD locus (see Supplementary Fig. 3 for details), while the wA locus remained unchanged. Ladder is Generuler 1 kb ladder (Thermo Scientific).
Fungal food production is a rapidly growing area with vast commercial interest and potential, and a growing number of meat and dairy substitutes based on fungi are now available on the market across Europe, U.S., and Asia 12 , 16 . While these products showcase the astonishing versatility and commercial promise of fungi for sustainable food production, most current products are based on a limited group of non-engineered strains, which have inherent limitations in their metabolism, structure, and industrial capacity. Genetic engineering could overcome these limitations and further expand beyond naturally occurring biodiversity, allowing new uses and applications of fungi in human food production 17 . For instance, a synthetic gene expression tool in Trichoderma reesei , an industrial fungus traditionally used for enzyme production, recently enabled production of gram-scale quantities of egg white and milk protein 14 , 18 . However, like many other industrial fungi, T. reesei has no history of safe or palatable consumption by humans, which limits the possible food contexts in which the fungus can be utilized, such as the highly efficient and sustainable production of fungal biomass for human food. Extending such synthetic biology tools and approaches to historically consumed, food-safe, edible fungi could expand the engineering possibilities for fungal food production, including enhancing fermented foods and altering the properties of mycoprotein to better suit human dietary needs and preferences. However, synthetic biology tools and demonstrated use cases for bioengineered food production by historically consumed food-safe filamentous fungi are lacking.
Here, we develop a modular synthetic biology toolkit for Aspergillus oryzae , an edible fungus with a long history of safe and palatable human consumption, and demonstrate its applicability for enhancing fungal foods. Our toolkit includes a CRISPR-Cas9 method for precise and efficient gene modification, neutral loci for targeted gene insertion, and tunable promoters, including bidirectional promoters as well as a synthetic expression system that offers strong gene expression independent of the medium composition. We use these tools to engineer the nutritional value and sensory appeal of the edible fungal biomass for alternative meat applications. We overproduce ergothioneine, a potent antioxidant, at levels that are higher than in mushrooms, the largest source of this molecule in the human diet. Additionally, we engineer the eight-step heme biosynthetic pathway to create an edible biomass that contains heme at levels approaching those found in leading plant-based meats incorporating heme for flavor and color. In contrast to plant-based protein, the engineered fungal biomass can be readily formulated into meat-like patties without the need for extensive processing, protein purification, or ingredient addition. In addition to demonstrating the potential of bioengineering edible fungi, this work provides synthetic biology tools and approaches that could be useful for fungi across diverse applications and industries.
A recyclable CRISPR-Cas9 method for efficient gene integration and expression
We selected Aspergillus oryzae (koji mold) as our model edible fungus and engineering target, as this fungus has a long history of safe use and human acceptance in fermented foods 13 , has a biomass with a palatable and umami-rich flavor that is commercially available as mycoprotein 19 , 20 , secretes high amounts of protein 21 , is used industrially for enzyme production 22 , and has demonstrated promise as a scaffold for animal cells in cellular agriculture 23 . To enable synthetic biology efforts across the diverse food applications of this edible fungus, we first set out to create a comprehensive genetic toolkit, including a method for efficient gene integration, neutral loci for high expression, and tunable promoters.
In designing our toolkit, we first considered the challenge of efficiently integrating heterologous genes in desired genomic locations. Filamentous fungi are notoriously poor at homology-based recombination and thus transformation with linear DNA templates often results in off-target, ectopic integrations 24 . To overcome this, strains deficient in non-homologous end-joining (NHEJ) have historically been used 25 . However, disruption of NHEJ presents potential issues with genomic instability or increased risk of DNA damage 26 . Recently, CRISPR-Cas9 has revolutionized the ability to transform and genetically modify fungi, including A. oryzae 18 , 27 , 28 , 29 . For example, the recently developed state-of-the-art CRISPR-Cas9 method for A. oryzae allows high-efficiency modification of strains proficient in non-homologous end-joining (NHEJ) 28 . The method uses plasmids to drive constitutive Cas9 and sgRNA expression, and the plasmid can be readily removed using selection, which enables sequential rounds of transformation and genome modification 28 .
To efficiently engineer A. oryzae , we sought to develop an alternative, easy-to-use CRISPR-Cas9 approach that is compatible with readily available commercial reagents 26 , minimizes the possibility of off-target effects and toxicity resulting from constitutive Cas9 expression 30 , 31 , 32 and incorporates a straight-forward phenotypic screen to verify that integration at the locus of interest has taken place 23 . Rather than encoding the Cas9 and sgRNAs from a plasmid, our method involves direct transformation of CRISPR-Cas9 Ribonucleoprotein complexes (RNPs), which can be formed in vitro from commercially available Cas9 protein and sgRNAs. At the start of our study, the RNP-based approach had been demonstrated as a strategy for rapid and precise genome editing compatible with high-throughput screening in diverse filamentous fungi 29 , 33 , 34 , 35 , 36 but had not been experimentally validated for gene integration in A. oryzae .
In our method, the DNA template introduced to fix the Double-Stranded Breaks (DSBs) harbors a pyrG marker to allow for both positive selection using uracil/uridine auxotroph and negative selection using media with 5-fluoroorotic acid (5-FOA). Although pyrG marker recycling has been demonstrated in A. oryzae , previous approaches did not incorporate CRISPR-Cas9 and required an NHEJ-deficient strain to avoid off-target integrations 37 . To overcome potential issues with ectopic integrations in wild-type A. oryzae , we designed the system such that a successful loop out of the pyrG marker can only occur if the fixing template is integrated at the locus of interest, in which case it will be flanked by two identical 300 bp sequences. In this system, ectopic integrations resulting from NHEJ are unable to loop out and survive on media supplemented with 5-FOA (Fig. 1B ). This phenotypic screening approach to locus-specific gene integration and pyrG marker recycling was first established using a plasmid-based CRISPR-Cas9 system in the related A. niger , where it allowed precise and efficient genome modification 27 . In A. niger , all colonies surviving on 5-FOA had the expected genome modification, highlighting the robustness of this approach. The A. niger method was used for mutation and gene deletion but was not utilized for integration and expression of proteins 27 .
To evaluate our RNP-based method for integration and expression in A. oryzae , we first targeted a GFP-expression cassette to the wA locus, which controls spore pigmentation, into a Δ pyrG mutant of the common laboratory strain RIB40 28 (Supplementary Table 1 ). This experiment yielded strains displaying the expected white spore phenotype, GFP expression, and fixing template insert (Fig. 1C, D , and Supplementary Fig. 1A, B ). To explore whether this method works beyond the model laboratory strain RIB40, we collected a group of A. oryzae strains with distinct industrial uses and geographical origins (Supplementary Fig. 2 ). Whole-genome sequencing of these strains revealed that these strains are phylogenetically distinct from one another and display minor variations in both the number of coding genes and biosynthetic gene clusters (Supplementary Table 2 ). To enable gene editing, we first generated Δ pyrG strains by targeting two RNPs to the pyrG locus and plating on agar supplemented with 5-FOA, uracil, and uridine to select mutants. PCR amplification of the region revealed clear mutations at the predicted sgRNA cut sites, including deletions and insertions, likely resulting from erroneous fixing by A. oryae . We observed no off-target effect on the surrounding genes despite not providing a fixing template, highlighting the precision of the RNP-based editing method (Supplementary Fig. 3 ). We then successfully introduced a GFP-expression cassette at the wA locus to alter the spore phenotype and establish heterologous protein production across the strain collection (Supplementary Fig. 4 ). These strains had not previously been genome sequenced or edited, suggesting that “wild” A. oryzae strains with potentially favorable phenotypes could be efficiently modified using our method. Moreover, these results indicate that the Δ pyrG mutants required for our method are readily generated in a single-step transformation.
The RNP-based CRISPR-Cas9 method was highly efficient. PCR amplification of RIB40 transformants at the wA locus revealed a 90% integration efficiency with 950 bp homology arms (Fig. 1E ), similar to the high targeting efficiency of the previously developed plasmid-based CRISPR-Cas9 system 28 . Even with homology arms as short as 25 bp, the method proved highly efficient (70%), suggesting that PCR primer overhangs could be used to specify the integration locus of interest (Fig. 1E ). The high efficiency with such small homology arms is consistent with findings from other filamentous fungi 34 , 38 . We also found that we could miniaturize the transformation to smaller volumes and remove the final top agar step without major decreases in integration efficiency, making the transformation process quicker and easier compared to the standard A. oryzae protoplast transformation protocol and compatible with a microplate format 28 , 35 (Supplementary Fig. 1E ). Although our method utilizes two RNP complexes for each locus to maximize the likelihood of DSB based on previous successes in fungi 27 , 39 , 40 , 41 , we found no major difference in integration efficiency between using one and two RNP complexes across two distinct genomic loci in A. oryzae ( wA and niaD ) (Supplementary Table 3 ).
A core design feature of our method is the ability to recycle the pyrG marker upon insertion to the correct locus, as transformants should undergo marker excision in the presence of 5-FOA. We first confirmed successful recycling from the wA locus. Consistent with previous results in A. niger 27 , surviving colonies displayed uracil-uridine auxotrophy and marker excision, indicating successful pyrG removal. We observed this across three out of three colonies analyzed, highlighting the robustness of the growth-based method to assess locus-specific marker recycling (Fig. 1F, G , and Supplementary Fig. 1C, D ). Finally, we successfully integrated GFP-expression cassettes and excised pyrG markers at the niaD locus, which controls nitrate assimilation, and the yA locus, which contributes to spore coloration (Supplementary Fig. 5 and Supplementary Table 1 ) 28 .
The recyclability of the pyrG marker allows for potentially endless rounds of sequential engineering. To evaluate this possibility, we transformed the looped-out wA : gfp strains with an mCherry cassette targeted to the niaD locus. Positive transformants demonstrated the expected phenotype and protein expression (Fig. 1H, I ). We then successfully recycled the pyrG marker from the niaD locus, enabling sequential engineering using our marker recycling approach (Fig. 1J ). Finally, we also targeted the wA and niaD loci simultaneously in a single experiment. However, in contrast to the high efficiency observed with single integration at the wA locus, simultaneous modification at the niaD and wA loci was only 30% efficient with 950 bp homology arms (Supplementary Fig. 6 ). This is consistent with previous findings of reduced efficiency with multiple RNP complexes and fixing templates in fungi 35 . Overall, these results establish the RNP-based method as a method for genome modification and protein expression in diverse strains of the edible fungus A. oryzae . This method displays a comparable high efficiency and scope as the plasmid-based method for genetically engineering this fungus 28 . The use of commercially available reagents and the ability to phenotypically screen for insertion at the locus of interest makes the protocol easy to use.
Identification and evaluation of neutral loci for gene expression
After establishing the RNP-based CRISPR-Cas9 method for gene modification, we considered another challenge in genetic engineering of filamentous fungi: where to integrate genes for overexpression. While multi-gene expression has been achieved in A. oryzae for natural products biosynthesis, the historically preferred method involves plasmids that integrate randomly throughout the genome 42 . These can cause unintended pleiotropic effects or genomic instability and make it challenging to compare phenotypes between constructs and strains 43 . In contrast, neutral loci, intergenic regions, and genomic safe havens that allow targeted expression without interfering with host physiology, is a standard feature of engineering for many bacteria and yeasts such as S. cerevisae 44 , 45 . Recently, genome sequencing of A. oryzae transformed with randomly integrated plasmids revealed two intergenic regions (called “hot-spots”) that were successfully targeted with CRISPR-Cas9 for expression of natural products genes 46 . However, to this date, neutral loci have not been systematically identified and evaluated for the efficiency of gene integration and level of protein expression across the A. oryzae genome. This information, along with readily available plasmids and DNA parts targeted to characterized loci, is critical to advance engineering efforts, as has been shown in S. cerevisae 45 .
We took a computational approach to identify candidate-neutral loci in A. oryzae . We first identified intergenic regions in the A. oryzae RIB40 genome. Using publicly available RNA-sequencing data across diverse conditions and growth stages, we ranked the expression level of the two genes immediately surrounding the intergenic region, thus generating a list of candidate loci predicted to enable high gene expression (Fig. 2A and Supplementary Data file 1 ). From this set, we selected 10 promising high-expression regions (>4.8 kb) spread across A. oryza e chromosomes for further evaluation (Fig. 2B , Supplementary Tables 4 and 5 ). We then integrated cassettes harboring GFP under control of the strong, constitutive pTEF1 promoter, and assessed fluorescence using flow cytometry on the conidia of looped-out strains 47 (Fig. 2B, C and Supplementary Fig. 7 ).
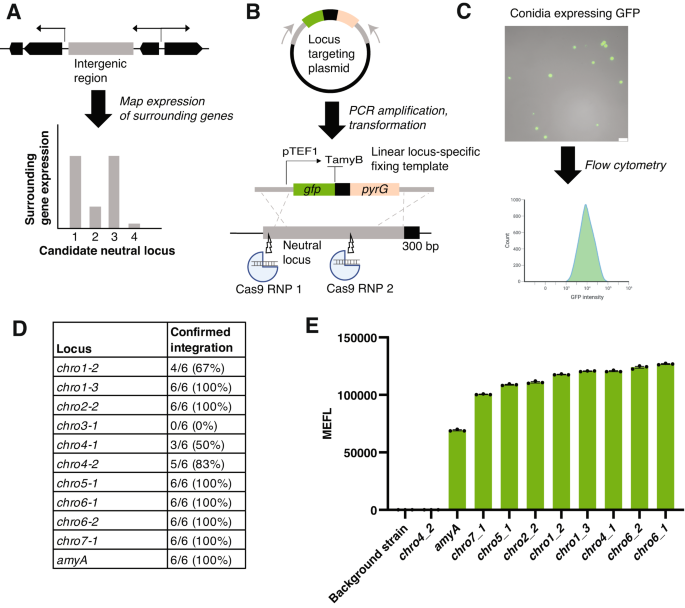
A A computational approach was used to identify intergenic regions with high expression of surrounding genes. The highest expressing regions were selected as promising neutral loci for further experimental evaluation. B Targeting plasmids were designed with the 5′ and 3′ homology arms as well as the specific 300 bp sequence for the locus of interest. The plasmid harbors a GFP-expression cassette driven by the constitutive pTEF1 promoter and terminated by the commonly used TamyB terminator, both from A. oryzae . The plasmids were cloned in E. coli and were linearized using PCR to create linear fixing templates that target the locus of interest. C Flow cytometry of conidia constitutively expressing pTEF1 was used to evaluate expression strength at the neutral locus of interest and determine their suitability for engineering efforts. A representative microscopy image showing GFP expression from A. oryzae conidia is shown. Scale bar = 25 µm. The flow cytometry figure was created was created using BioRender ( http://BioRender.com ). D Integration efficiency of GFP-expression cassette at neutral loci. All loci except for chro3_1 displayed a high efficiency of integration (>50%), as assessed by PCR. We could not detect insertion at chro3_1 by PCR. E GFP expression (expressed as Mean Equivalents of Fluorescein, or MEFL) across neutral loci. All loci except for chro4_2 displayed expression levels above the background strain (RIB40) and were higher than the amyA locus, which was included as a positive control to validate the method. Results are average and standard error of the mean (SEM) of three biological replicates.
Out of 10 tested loci, 9 showed high-efficiency integration (>50%) (Fig. 2D ). We could not detect successful gene insertion at chro3_1 by PCR amplification, suggesting issues with PCR amplification or the integration itself. Following marker loop out, we detected GFP expression above background levels from 8 of the remaining 9 loci, with chro4_2 displaying no clear GFP expression (Fig. 2E ). Expression levels were largely consistent across the loci, with the highest (chro6-1) showing ~25% higher expression than the lowest (chro7-1) (Fig. 2E ). All loci displayed higher expression than the amyA locus, which was included as a positive control to validate the method. Finally, colony growth and morphology were consistent between all strains and similar to those of the background strain, suggesting no gross effects of gene integration and expression on fungal growth (Supplementary Fig. 8 ). Overall, these efforts identified not only neutral loci, but also a set of plasmids and sgRNAs that facilitates easy transformation and expression for diverse purposes (Supplementary Table 5 ). Our computational identification and experimental evaluation using flow cytometry provides a framework for how to identify and evaluate promising candidate loci across fungal hosts.
Expansion of the promoter toolkit using a synthetic expression system and bidirectional promoters
An additional challenge in engineering edible filamentous fungi is the narrow set of characterized parts available for gene regulation, as fungal promoters remain limited in both sophistication and scope. For example, only a handful of endogenous promoters have been used for gene expression in A. oryzae , and these are either regulated by the nutrient source (such as the amylase or glucoamylase promoters), have a limited dynamic range, or a poorly understood mode of regulation 48 , 49 , 50 . Synthetic expression systems (SES), which are widely available in yeast and bacteria and increasingly in mammalian cells and plants, could address the technical limitations of current fungal expression tools and expand engineering opportunities in A. oryzae 51 , 52 , 53 , 54 , 55 , 56 , 57 . SES couple synthetic transcription factors (sTF) with minimal core promoters (Cp) and DNA binding sites (UAS) and offer an orthogonal and highly programmable mode of gene expression 18 (Fig. 3A ). The Tet-On SES has shown promise in A. niger and A. fumigatus for inducible and titratable gene expression 58 , 59 , but food applications are limited by the cost and potential food incompatibility of the small molecule inducer. In contrast, a constitutive SES based on the Bm3R1 DNA binding domain and the VP16 activation domain was recently established in the two non-edible, industrial filamentous fungi A. niger and T. reesei . The highly modular SES showed high programmability and stability across the two hosts and afforded high secreted protein expression independent of the composition of the growth medium 18 .
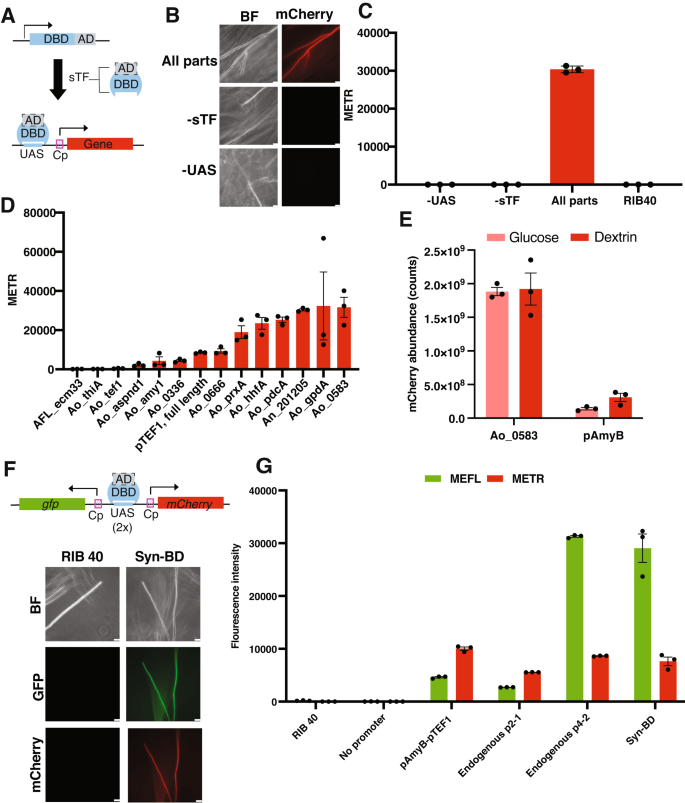
A Design of synthetic expression system (SES). Coupling a synthetic transcription factor (sTF, composed of an Activating Domain = AD and DNA binding domain = DBD) and Upstream Activating Sequences (UAS) enables orthogonal and highly programmable gene expression from core promoters (Cp). B Confirmation of the Bm3R1-VP16-based SES in A. oryzae using the An_201205 core promoter. Fluorescence imaging that the SES in A. oryzae requires both the sTF and the UAS for expression. Scale bar = 50 µm for −UAS, 25 µm for other strains. C Conidia from strains shown in ( B ) were subjected to flow cytometry for fluorescence quantification of the constitutively expressed mCherry (expressed as Mean Equivalents of Texas Red, or METR). Results are average and SEM of three biological replicates. D Core promoter screen using the SES in A. oryzae . 200-bp sequences were cloned upstream of mCherry and fluorescence intensity was quantified using flow cytometry of conidia. The full-length, constitutively expressed promoter pTEF1 was included as a benchmark for promoter strength. Results are average and SEM of three biological replicates. E Proteomic comparison of intracellular mCherry abundance between the core promoter, Ao_0583, and the full-length starch-inducible endogenous promoter pAmyB from A. oryzae . Proteomics was conducted on lyophilized mycelia grown in liquid cultures (CDA medium with dextrin or glucose as the sole carbon source). Results are average and SEM of three biological replicates. F A minimal bidirectional promoter (Syn-BD) constructed of 2x UAS binding sites and the gpdA and hhfA core promoters can drive dual mCherry and GFP expression. Scale bar = 25 µm for RIB40, 50 µm for engineered strain. G Identification and evaluation of endogenous bidirectional promoters from A. oryzae . Flow cytometry quantification indicated that two promoters, p2-1 and p4-2, could drive bidirectional gene expression at varying levels. p4-2 was similar to Syn-BD in terms of expression. A concatenated sequence of pAmyB and pTEF1 pointing in opposite directions was included as a positive control. MEFL = mean equivalents of fluorescein. METR = mean equivalents of Texas red. Results are average and SEM of three biological replicates.
We sought to expand the engineering possibilities in the edible A. oryzae by building on these advances in the industrial workhorses A. niger and T. reesei . To first establish SES as a mode for gene regulation in A. oryzae , we initially evaluated the ability of the previously characterized Bm3R1-NLS-VP16 sTF to drive mCherry expression from a core promoter. Using the RNP-based CRISPR-Cas9 integration tools and neutral loci, we genetically integrated the sTF and drove low levels of basal expression of this transcription factor using a characterized core promoter from A. niger (An008). In a separate genomic location, we integrated an mCherry cassette harboring 6x UAS upstream of the A. niger An201205 core promoter. There was clear expression of mCherry in mycelia and conidia using the full system. The UAS and the sTF were both necessary for activity, validating the predicted function of the SES in A. oryzae 18 (Fig. 3B, C ).
To explore the programmability of this modular SES in A. oryzae , we initially focused on core promoters, as the identity of these short 200-bp sequences influences the level of gene expression upon sTF binding 18 , 57 . Using available transcriptome data and a curated list of promoters from highly expressed A. oryzae and Aspergillus flavus genes, we first assembled an initial library of twelve 200-bp core promoters and evaluated their ability to drive mCherry expression in the SES (Supplementary Table 6 ). We used flow cytometry of conidia 47 as an initial screening approach to assess expression and used the strong, constitutive pTEF1 promoter as a benchmark for comparison. Three of twelve selected core promoters did not drive mCherry expression at levels above the background strain (Fig. 3D ). However, across strains producing detectable mCherry, mean expression across the core promoter library displayed a 14-fold expression range, from 0.25 to more than 5-fold pTEF1. These results indicate that, like full-length promoters, core promoter sequences can drive divergent transcriptional outputs in A. oryzae (Fig. 3D ). Proteomics analysis of mCherry in biomass grown in submerged fermentations confirmed that core promoters could also drive protein expression in mycelia, including at levels that were several-fold higher than pTEF1 (Supplementary Fig. 9A ). There was a significant correlation between the flow cytometry and proteomics data ( r = 0.82, R 2 = 0.67, p < 0.01, Supplementary Fig. 9B ). This suggests that flow cytometry is a useful screening approach to identify constitutive promoters. Nonetheless, following up on flow cytometry screening results in mycelia may be needed for establishing the precise promoter strength for submerged fermentations.
To further benchmark the SES system we used proteomics to compare the strength of the SES promoter Ao_0583, identified as >4-fold stronger than pTEF1 in both conidia and mycelia, with the starch-inducible pAmyB promoter, one of the strongest known endogenous A. oryzae promoters that is frequently used for high protein expression and secretion from submerged cultures 21 , 50 . Strikingly, Ao_0583 was approximately 6-fold stronger than that of the pAmyB promoter when the strain harboring it was grown under inducing conditions. mCherry levels were estimated to comprise approximately 13% intracellular protein under the Ao_0583 system, and only 1–2% in the pAmyB expression strain (Fig. 3E and Supplementary Fig. 10 ). While mCherry levels did not differ between glucose and dextrin when using the SES, pAmyB expression increased on dextrin, the predicted inducer of pAmyB (Fig. 3E and Supplementary Fig. 10 ). Thus, the SES permits high protein expression independently of the carbon source and avoids the complex multi-step regulation and global metabolic changes involved in pAmyB-driven expression in A. oryzae 60 . To our knowledge, the expression levels afforded by the SES far outperform any characterized promoter in A. oryzae .
In addition to the limited set of available mono-directional promoters, there is a lack of bidirectional promoters for filamentous fungi. Bidirectional promoters, which are available in yeast, could accelerate genetic engineering in edible filamentous fungi by enabling assembly of multi-step metabolic pathways or multi-protein complexes through fewer transformations 61 , 62 . We addressed this challenge in two ways. First, we created a synthetic bidirectional promoter (Syn-BD), as the modular nature of the SES enables different parts to be combined to create highly programmable modes of gene expression 63 . By combining two core promoters ( gpdA and hhfA ) with 2× UAS binding sites, we created a 485-bp bidirectional promoter was sufficient to drive bidirectional gene expression using the SES (Fig. 3F ). Second, we computationally identified candidate endogenous bidirectional promoters using publicly available RNAseq data collected across diverse growth conditions (Supplementary Data file 2 ). Out of five computationally identified bidirectional promoter candidates (Supplementary Table 7 ), two (p2-1 and p4-2) could drive mCherry and GFP expression in a bidirectional fashion (Supplementary Fig. 11 ). The p4-2 promoter displayed similar levels of expression as the rationally designed Syn-BD and a control bidirectional promoter composed of concatenated pAmyB-pTEF1 sequences pointing in separate directions (Fig. 3G ). Interestingly, the sequence of the identified p4-2 endogenous promoter in A. oryzae has the same length and genomic context as the H3/H4 histone promoter which was previously identified and evaluated in a range of Aspergilli , but not in A. oryzae 64 . This suggests that our computational pipeline might be broadly useful to identify bidirectional promoters in filamentous fungi. Overall, these results expand the set of gene regulation tools and promoters available for engineering the edible A. oryzae .
Edible mycelium bioengineered for enhanced nutritional value and sensory appeal
Having established a synthetic biology toolkit for A. oryzae , we next sought to deploy our tools to bioengineer its edible mycelium, as a first step toward enhancing its value as mycoprotein. We were inspired by the recent commercial success of bioengineered Saccharomyces used for brewing, which have been modified for improved cost savings, sustainability, and sensory profiles 17 . To explore whether genetic modification could similarly enhance foods made with filamentous fungi, we set out to modify endogenous biosynthetic pathways that could potentially improve the nutritional value and sensory appeal of the edible A. oryzae mycelium for alternative meat applications.
We initially focused our engineering efforts on ergothioneine, a bioactive amino acid and powerful antioxidant. Low plasma levels of ergothioneine are correlated with cardiovascular disease and neurological decline, and humans encode a specific ergothioneine transporter that uptakes ergothioneine from the diet, underscoring the potential importance of this molecule in human health 65 . While many foods contain low levels of ergothioneine, fungi are the major dietary source 66 . Work in the model ascomycete mold Neurospora crassa has revealed that fungal ergothioneine biosynthesis involves two enzymes, Egt1 and Egt2, that convert cysteine, S-adenosylmethionine, and histidine, to ergothioneine 67 , 68 , 69 (Fig. 4A ). N. crassa Egt1 and Egt2 were recently co-expressed in A. oryzae using plasmid-based random integration 70 . Rice cultured with transformants in solid-state fermentation had elevated ergothioneine levels, but the levels in the biomass alone were not investigated. Untransformed A. oryzae produced low levels of ergothioneine, suggesting that this fungus may harbor endogenous pathways for production 70 .
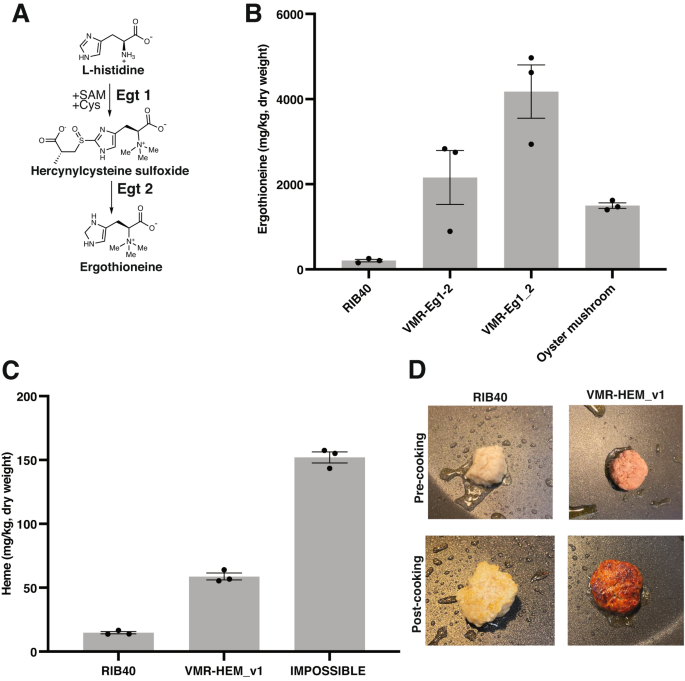
A Fungal biosynthesis of ergothioneine, a powerful antioxidant associated with several health benefits in humans. The characterized biosynthetic pathway from the fungus Neurospora crassa involves the enzymes Egt1 and Egt2. B A. oryzae homologs of N. crassa Egt1 and Egt2 were identified bioinformatically (see Supplementary Table 7 and Supplementary Fig. 12 for details) and expressed from neutral loci using a bidirectional promoter (strain VMR-Eg1-2) or as two separate genes at two different genomic locations, with each gene under the control of its own promoter (strain VMR-Eg1_2). The strategy is described in Supplementary Fig. 12 . Oyster mushroom, the dietary mushroom with the highest ergothioneine content, was included for comparison. Biomass was analyzed by LC–MS. Results are average and SEM of three biological replicates. C Engineering of heme biosynthesis in A. oryzae biomass. The strategy is described in Supplementary Fig. 15 . Heme was quantified using LC–MS in the biomass. The intracellular heme levels in the engineered strain were 4-fold higher than in the background strain, RIB40, and 40% of those found in IMPOSSIBLE™ burger made from plants, a leading plant-based meat product incorporating heme for flavor and color, was included for comparison. Results are average and SEM of three biological replicates. D Color of harvested background and engineered heme strain after culturing. The engineered strain overproducing heme (VMR-HEM_v1) was distinctly red in color, while RIB40 was off-white. The harvested fungal biomass could be readily formulated into an imitation meat patty with minimal processing. The color difference remained upon cooking, further enhancing the meat-like appearance of the naturally textured fibrous biomass.
Instead of introducing foreign genes, we hypothesized that by changing the expression of potential endogenous A. oryzae genes involved in ergothioneine biosynthesis, we could elevate production in the edible biomass to levels found in dietary mushrooms. To identify candidates, we searched the A. oryzae genome for homologs of N. crassa Egt1 and Egt2. We found two A. oryzae ortholog candidates sharing 49.2 and 45.1% amino acid sequence (Supplementary Table 8 ). Sequence alignment indicated conservation of key residues or functional groups bioinformatically predicted to be involved in substrate binding in Egt1, as well as residues structurally confirmed to participate in catalysis in Egt2 68 (Supplementary Fig. 12 ). We then integrated the A. oryzae homologs (named AO_Egt1 and AO_Egt2) at neutral loci and drove expression of both genes from either a bidirectional promoter (strain VMR-Eg1-2), or as two separate genes in two separate genomic locations (strain VMR-Eg1_2) (Supplementary Fig. 12 ). High-resolution Liquid Chromatography-Mass Spectrometry (LC–MS) was used to detect ergothioneine in samples (Supplementary Fig. 13 ). Consistent with previous observations 70 , we detected low levels of ergothioneine in the mycelium in the background strain RIB40 (Fig. 4B and Supplementary Fig. 13 ). However, the bidirectional promoter and separate promoter strains elevated ergothioneine 11-fold and 21-fold, respectively, over RIB40 (Fig. 4B ). While the ergothioneine levels in VMR-Eg1-2 was similar to those found in oyster mushroom, the highest known dietary ergothioneine source 66 , the mean levels in strain VMR-Eg1_2 were 1.5-fold higher. We observed no major difference in protein content between the wild-type and engineered strains; however, ergothioneine overproduction was associated with a slight growth defect, suggesting a metabolic burden of ergothioneine production under the growth conditions (Supplementary Fig. 14 ). Overall, these results implicate the endogenous genes AO_Egt1 and AO_Egt2 in ergothioneine biosynthesis in A. oryzae and validate the metabolic engineering approach to alter the molecular composition of mycoprotein.
Having validated our tools to increase levels of bioactive molecules for enhanced nutritional value, we asked whether a similar approach could be applied to sensory properties of the edible biomass to more closely mimic animal meat. For example, even though the A. oryzae biomass has a meat-like fibrous texture owing to its microscopic morphology, the biomass, which is off-white, would necessitate color addition for many meat applications. As a first step toward improving the meat-like flavor composition and appearance of the edible biomass using bioengineering, we initially targeted the biosynthesis of heme, an essential cofactor that catalyzes a wide range of reactions across all domains of life and gives red meat its color and contributes to flavor upon cooking 71 . IMPOSSIBLE Foods, a leading plant-based meat producer, has taken advantage of these properties of heme and adds a purified soy Leghemoglobin (LegH) produced with the yeast Pichia pastoris to its products based on plant protein isolates to create realistic alternatives that look like red meat 72 , 73 . Other plant-based meat producers have now followed suit with similar hemoglobin addition strategies 74 .
We reasoned that by modulating the expression of key heme biosynthetic enzymes, we could elevate intracellular heme in the edible fungal biomass to levels found in leading meat alternatives incorporating heme for flavor and color. Fungal heme biosynthesis is carefully regulated at the transcriptional and post-translational levels and involves eight dedicated enzymes, which are split between the mitochondria and the cytosol 75 (Supplementary Fig. 15 ). We identified potential heme biosynthesis proteins in A. oryzae by searching the genome for sequences found in S. cerevisiae 76 (Supplementary Table 9 ). There is limited experimental information about heme biosynthesis in filamentous fungi, but based on successful engineering efforts from yeast 77 , 78 , and studies of individual heme biosynthetic enzymes in different Aspergilli 75 , 79 , 80 , 81 , we initially targeted expression of predicted rate-limiting enzymes, including ALAS (biosynthetic enzyme#1), PBGD (#3), UROD (#4), and CPO (#5). Additionally, we mutated key cysteine residues in the Heme Regulatory Motif (HRM) of ALAS to remove potential feedback inhibition by heme 82 (Supplementary Fig. 15 ). Importantly, high levels of free heme and the porphyrin intermediates can be toxic to the cell, causing oxidative damage and hampering growth 77 , 83 . To address this potential challenge, we expressed two copies of Soy Leghemoglobin, the FDA-approved protein used in IMPOSSIBLE meat 72 , as a potential heme sink, using both the SES and the significantly weaker pTEF1 promoter. Though the regulation of heme biosynthesis has not been characterized in detail in filamentous fungi, simultaneous elevation of biosynthetic enzymes and a heme-binding protein was necessary to increase heme levels without causing excessive toxicity in S. cerevisiae 77 , 78 . The final engineered strain A. oryzae contained a total of separate six modifications (Supplementary Fig. 15 ).
We used high-resolution LC–MS to detect heme across all samples (Supplementary Fig. 16 ). The biomass of the engineered strain contained 4-fold higher levels of heme compared to the non-engineered strain, on a dry weight basis. These levels of heme in the engineered strain were nearly half (40%) of those found in IMPOSSIBLE meat (Fig. 4C ). Increasing levels further may require tuning pathway flux or making additional modifications beyond biosynthetic enzyme expression levels, as was recently shown in S. cerevisae 76 . However, to our knowledge, this is the highest levels of intracellular heme in fungal mycelium and a rare example of heme biosynthesis engineering in filamentous fungi. Given the importance of heme for enzyme production for biofuels and medical applications 78 , 84 , we envision that these strains and approaches could have broad applicability for engineering efforts beyond food.
Upon harvesting the biomass of the heme overproducer, we noticed that it was red in color, compared to the off-white color of the background strain. In contrast to other plant-based meat alternatives, which require extensive processing and ingredient addition to transform off-flavor plant protein isolates (such as soy or pea) to meat alternatives, this bioengineered mycoprotein required minimal post-harvest processing for formulation into an imitation red meat patty following a standard mycoprotein production protocol 11 (Fig. 4D ). The only processing needed was removing excess liquid from the biomass prior to grinding and cooking. The color difference between the background and engineered strains remained after cooking, enhancing the meat-like appearance of the naturally textured, fibrous fungal biomass (Fig. 4D ). There was no decrease in the growth yield or protein content (46%, on a dry weight basis) in the engineered heme strain relative to the background strain (Supplementary Fig. 17 ). The engineered mycoprotein also contained all the essential amino acids, suggesting a promising nutritional profile (Supplementary Fig. 17 ). Taken together, these data suggest that the engineered edible fungal mycelium could have promise in meat alternative applications.
Filamentous fungi are widely used for the industrial production of enzymes and metabolites and recently have found more widespread use in both sustainable materials and foods 12 . However, genetic tools for these organisms have historically been limited in both sophistication and scope, preventing both engineering efforts and fundamental studies. Recent advances in CRISPR-Cas9 technology have dramatically improved the possibilities of modifying diverse mushrooms and molds, including the food-safe, edible fungus A. oryzae 27 , 28 , 29 , 34 . In contrast to industrial strains such as T. reesei , which was recently used to produce milk and egg proteins at lab scale 14 , the historically consumed A. oryzae has potential uses across fermented foods, food protein production, cellular agriculture, and mycoprotein 13 , 20 , 22 , 23 .
To enable bioengineering for these diverse applications, we developed a ready-to-use toolkit that is now available to the research community and includes DNA parts for integration and regulation of genes and pathways. Similar toolkits are available in S. cerevisiae , where they have significantly expanded opportunities for genetic engineering 45 . We hope that our tools will be similarly useful for expanding the engineering possibilities in A. oryzae , alongside other recently developed genome modification methods such as base editing 85 , in vivo DNA assembly in NHEJ-deficient strains 86 , and protein expression screening 87 . Additionally, we envision that the computational and experimental approaches used here – for identification and evaluation of neutral loci and design and identification of promoters – could be broadly useful for constructing genetic toolkits and engineering diverse fungal hosts.
We used our tools to enhance the molecular composition and appearance of the mycelium as a first step toward improving its nutritional and sensory properties. First, we engineered A. oryzae mycoprotein to overproduce the nutraceutical ergothioneine at levels that are higher than those in mushrooms, the highest known natural source from the diet. While ergothioneine has been produced in a range of microbial hosts for the purpose of isolating the nutraceutical 88 , 89 , our work represents a proof of concept of modifying endogenous ergothioneine biosynthesis for mycoprotein applications. Separately, we engineered the A. oryzae mycelium to overproduce heme, a key flavor-and-color molecule in red meat, at levels that are close to half those found in leading plant-based meats. Our engineering of the edible fungal biomass for alternative meat presents an alternative approach to fungal food beyond the production of secreted animal proteins, which is a less efficient fermentation process and has a higher environmental footprint than biomass production 6 , 9 . Future engineering targets for edible fungal biomass could include lipid pathways for flavor, amino acids for nutrition, structural alteration for texture improvement, or enzymes for improved growth on affordable, complex feedstocks. However, it is important to note that our work represents early prototypes, and further assessment of the sensory attributes, consumer acceptability, potential food safety concerns, and the regulatory landscape around genetically modified organisms (GMO), is needed to bring engineered edible fungi from lab bench to the table.
A. oryzae , like many of the strains that form the basis of fungal foods available on the market, has been genetically modified through extensive selection and breeding throughout human history 90 , 91 . Genetic modification using contemporary gene editing tools such as CRISPR-Cas9 represents a natural next step in this long history of microbial gene modification to suit human needs and holds promise to further expand fungal strain diversity and accelerate the adaptation of fungal strains to the demands of current production methods and consumer preferences. Bioengineered edible plants and yeasts have demonstrated reduced environmental impact, improved nutrition, and improved flavor profiles compared to their non-engineered counterparts and are already available on the market 17 , 92 , 93 . We anticipate similar possibilities with genetic modification of edible filamentous fungi, as synthetic biology in these organisms is uniquely positioned to address the pressing environmental, ethical, and public health challenges of industrial animal agriculture.
All primers used for genome modification are shown in Supplementary Table 10 . All strains and plasmids used for strain construction are listed and described in Supplementary Tables 11 - 12 . The sequence files corresponding to each strain and plasmid can be found in the JBEI Public Registry ( https://public-registry.jbei.org/ ) 94 . All plasmids were propagated in Escherichia coli strain DH10B and purified by Miniprep (Qiagen). The plasmids generated in this study were based on the pTWIST_amp backbone (TWIST biosciences) and were constructed by Gibson assembly 95 using Gibson assembly master mix (New England Biolabs). PCR amplification was performed using NEB Q5 polymerase according to the manufacturer’s instructions (New England Biolabs). All genes were codon optimized for A. oryzae and ordered either as G-blocks from IDT or as complete, sequence-verified genes from IDT or TWIST biosciences. The coding sequences of heterologous genes in all plasmids were validated by Sanger sequencing (Azenta) or whole-plasmid sequencing (Primordium).
Growth conditions
A. oryzae strains were always grown at 30 °C. A variety of media were used in the transformation and cultivation of A. oryzae , and these are indicated below. They are referenced in the materials and methods section. Media were supplemented with 5 g/L uridine (Sigma–Aldrich, #U6381) or 2 g/L uracil (Sigma–Aldrich, #U1128) when supplementation to support growth of pyrG mutants was needed. Supplementation is indicated as UU throughout the manuscript.
GP medium (per 1 L of medium)
5 g yeast extract, 10 g polypeptone, 0.5 g MgSO 4 ⋅ 7H 2 O, 5 g KH 2 PO 4 . 20 g glucose was used as the carbon source unless otherwise indicated. Alternatively, 20 g dextrin (Sigma–Aldrich, #31400) was used as the carbon source.
PDA + 5-FOA + Uridine + Uracil (PDA + 5-FOA + UU) medium (per 1 L of medium)
39 g Potato Dextrose Agar (PDA, Sigma–Aldrich, #70139-500 G), 5 g uridine,2 g uracil, and 1 mg/mL 5-fluoroorotic acid (ThermoFisher, #R0812).
Bottom Agar + Methionine (BA + Met, per 0.5 L of medium)
1 g NH 4 Cl, 0.5 g (NH 4 ) 2 SO 4 , 0.25 g KCl, 0.25 g NaCl, 0.5 g KH 2 PO 4 , 0.25 g MgSO 4 •7H 2 O, 0.01 g FeSO 4 , 109.3 g sorbitol, 7.5 g agar, 10 g glucose, 0.75 g methionine. pH was adjusted to 5.5 prior to autoclaving.
Top Agar + Methionine (TA + Met, per 0.5 L of medium)
Same as BA + Met, but 4 g agar instead of 7.5 g agar per 0.5 L.
Minimal Medium Agar + Methionine (MMA + Met, per 0.5 L of medium)
Same as BA + Met, but no sorbitol added as the osmotic stabilizer.
CDA medium (per 1 L of medium)
3 g NaNO 3, 2 g KCl, 1 g KH 2 PO 4 , 0.5 g MgSO 4 × 7 H 2 O, 0.02 g FeSO 4 ·7H 2 O, 15 g agar. 20 g glucose was used as the carbon source unless otherwise indicated. Alternatively, 20 g dextrin (Sigma–Aldrich, #31400) was used as the carbon source.
CDA(Leu) medium
same as CDA medium but containing 10 mM leucine as the sole nitrogen source instead of the 3 g/L NaNO 3 .
Strain construction
A. oryzae strains were genetically modified using protoplast transformation (see standard transformation protocol below). All A. oryzae strains are described in Supplementary Table 12 . Regenerated protoplasts were restreaked onto MMA + Met plates to obtain single colonies and purify the potentially heterokaryotic conidia. Following 48 h of growth at 30 °C, the conidia of individual, single colonies were transferred to MMA + Met slants for growth for 48–72 h at 30 °C. These purified strains represented the strains used in all assays and characterizations. To confirm the insertion of genes at the correct locus, colony PCR was performed on conidia on slants using PHIRE direct plant PCR kit (ThermoFisher, #F130WH) by boiling conidia in 20 µL of dilution buffer for 10 min at 95 °C and using 1 µL of the conidial spore suspension as the template for PCR, which was set up according to the manufacturer’s instructions. Strains harboring the correct insertions were saved as glycerol stocks by suspending conidia in 30% glycerol (v/v). For the simultaneous targeting of wA and niaD loci in a single transformation, DNA templates of plasmids were prepared as described below and 10 µg of each plasmid, along 5 µL of each RNP complex (four total, two per locus) were added at the DNA-RNP incubation step. To check for spore coloration ( wA and yA mutants), strains were grown on PDA medium for 5 days at 30 °C. To check for nitrate assimilation ( niaD mutant), strains were grown on CDA and CDA-Leu for 5 days at 30 °C. To check for pyrG mutation and the associated uridine/uracil auxotrophy, strains were grown on CDA and CDA supplemented with 2 g/L uracil and 5 g/L uridine for 5 days at 30 °C. To assess the targeting efficiency at individual loci, and to evaluate the effect of homology arm length on integration at the wA locus, colony PCR of 10 individual strains was performed, and those displaying the correct band by PCR were deemed successful integrations. Varying homology arm lengths of the wA fixing template were obtained by linearizing the full-length template with different primers (see primer table, Supplementary Table 10 ). Flow cytometry or microscopy assays were used to assess the expression of fluorescent proteins (see below for details). To create the engineered strain VMR-Eg1-2, A. oryzae RIB40 pyrG mutant was transformed with the linear DNA template originating from JBx_250940 and the two 5′ and 3′ RNP complexes targeting the chro1-3 neutral locus. To create the engineered strain VMR-Eg1_2, A. oryzae RIB40 pyrG mutant was transformed with the linear DNA template originating from JBx_250936 and the two 5′ and 3′ RNP complexes targeting the chro1-3 neutral locus, and subsequently with the linear DNA template originating from JBx_250938 and the two 5′ and 3′ RNP complexes targeting the chro2-2 neutral locus. For the engineered strain overproducing heme (VMR-HEM_v1), A. oryzae RIB40 pyrG was sequentially transformed with linearized DNA originating from plasmids JBx_250942, JBx_250944, JBx_250946, JBx_250948, JBx_250950, JBx_236225, as well as the specific 5′ and 3′ RNP complexes associated with the target locus for integration. For the cultivation of RIB40, and the engineered strains overproducing ergothioneine and heme, 5 × 10 5 conidia were inoculated into 50 mL of GP-glucose medium (ergothioneine strains and corresponding RIB40 control) or 50 mL GP-dextrin medium (heme strain and corresponding RIB40 control) in 250 mL Erlenmeyer flasks. The strains were grown for 96 h at 30 °C, shaking at 160 rpm. Biomass was harvested by vacuum filtration over Miracloth. Biomass was lyophilized for extraction of metabolites and was dried for 7 days at 50 °C prior to recording of the dry mass.
Transformation of A. oryzae
Preparation of linearized dna fixing templates for transformation.
To generate linear DNA to be transformed into A. oryzae as fixing templates alongside CRISPR-Cas9 RNP complexes, the DNA was linearized using PCR from the corresponding plasmids harboring the DNA fixing template of interest. Briefly, 1 ng of plasmid DNA was used as the template for a 60 µL PCR reaction using the Q5 high-fidelity polymerase master mix (New England Biolabs, #M0492S) and following the manufacturer’s instructions for the PCR protocol. For each DNA template, five 60 µL reactions were set up in parallel to obtain sufficient DNA for transformation. The PCR reactions were then combined and purified using the QIAquick PCR purification kit (Qiagen) and were eluted at the final step in 25 µL of sterile water. This typically gave sufficient quantities of the large amount of DNA needed for protoplast transformation (>10 µg in 20 µL).
Preparation of RNP complexes for transformation
All CRISPR-Cas9 reagents were obtained from IDT, including the Alt-R S.p. HiFi Cas9 Nuclease V3 (IDT, #1081061), Alt-R CRISPR-Cas9 crRNA XT 2 nmol (customized sequence), and Alt-R CRISPR-Cas9 tracrRNA 100 nmol (IDT, #1072534). RNA duplex buffer was included as part of the tracrRNA. The crRNA, which is the sequence-specific RNA that targets the region of interest, was resuspended in 20 µL water for 100 µM final concentration. To hybridize the sequence-specific crRNA to the universal tracrRNA to generate the final sgRNA, 0.5 µL of crRNA (5 µM final concentration) was mixed with 0.5 µL tracrRNA (5 µM final concentration) in 9 µL RNA duplex buffer and the mixture was then heated for 5 min at 95 °C and was then left to cool. This hybridized mixture represents the final sgRNA which is ready to bind to the Cas9 protein to form the RNP complex. To create the final RNP complexes for transformation, 2.16 µL of the hybridized crRNA-tracrRNA (the sgRNA, final concentration 540 nM) was mixed with 0.18 µL of Alt-R S.p. HiFi Cas9 Nuclease V3 (final concentration 540 nM) and 17.66 µL buffer (9 mM HEPES, 67 mM KCl, pH 7.5). The mixture was incubated at room temperature for 20 min to form the RNP complex and was then transferred to ice. As specified below in the transformation protocol, 5 µL of each RNP complex was used per 200 µL protoplast transformation. The RNP complexes were always prepared fresh on the day of the transformation and were never subjected to freeze-thaw.
Standard transformation protocol
Protoplast-mediated transformation was used to transform Aspergillus oryzae . We followed the protocol from 28 , with minor modifications. To generate mycelial biomass for protoplast generation, pyrG mutant strains were grown in duplicate in 50 mL GP medium supplemented with uracil and uridine in 250 mL flasks at 30 °C, shaking at 160 rpm. Following 72 h of growth, mycelia were harvested by pressing the liquid from the mycelia in a 20 mL syringe harboring a sterile cotton ball.
Dry mycelia from one flask were then put in 10 mL of TF1 solution (Per 500 mL of water: 2.9 g maleic acid, 39.5 g (NH 4 ) 2 SO 4 , pH adjusted to 5.5 and filter sterilized) harboring 0.1 grams of YATALASE enzyme (Takara Bio, #T017) which was used to digest the cell wall. This was incubated shaking for two and a half hours at 30 °C and 160 RPM, and the tissue was pressed within a sterile syringe with cotton to collect the protoplasts within the flow-through. The flow-through was checked for cloudiness, which indicated the generation of protoplasts. The resulting protoplast solution (7–8 mL usually, as sometimes not all protoplast solution could be pushed through the cotton ball due to clogging) was centrifuged at 475 g for 10 min, and the supernatant was then discarded. The protoplasts were then gently resuspended in 10 mL of TF2 solution pre-warmed at 30 °C (Per 1 L of water: 218.5 g sorbitol, 10.95 g CaCl 2 •6H 2 O, 2.05 g NaCl, 1.21 g Tris buffer, pH adjusted to 7.5 and filter sterilized). The protoplast solution resuspended in TF2 was centrifuged at 475 g for 10 min to discard the supernatant. The protoplasts were resuspended in 1–2 mL of TF2 solution and 200 μL of protoplast solution was placed into 15 mL volume centrifuge tubes. At least 1.2 × 10 7 protoplasts/mL was needed, so it is useful to consider this concentration when resuspending in the 1–2 mL TF2 solution. Concentrations of ~10 8 protoplasts per mL were typically obtained in the standard digestion protocol.
To transform the protoplasts, a total of 10 µg of PCR-linearized DNA fixing template (in 20 µL sterile water) was added to the 200 μL protoplast solution. Then 5 µL of each of the two pre-formed sgRNA-Cas9 RNP complexes were added and the protoplast-DNA-RNP mixture was incubated on ice for 30 min. After 30 min, sequentially and slowly, 250 μL, 250 μL, and 850 μL aliquots of TF3 solution were added (TF3 solution: per 1 L of water: 600 g Polyethylene glycol (4000), 10.95 g CaCl 2 •6H 2 O, 1.21 g Tris buffer, pH adjusted to 7.5 followed by autoclaving) and left to incubate at room temperature for 30 min. A total of 5 mL of TF2 solution was then added to each protoplast solution then centrifuged for 10 min at 475 g to discard the supernatant. The protoplast pellet was resuspended in 500 μL of TF2. At this point, a bottom agar plate pre-warmed at 30 °C already brought out from its heating location (see above for bottom agar recipe). The 500 μL of protoplast suspension was then mixed with 5 mL of a liquid layer of top agar (see above for recipe) pre-warmed to 50 °C, then quickly spread uniformly across the bottom agar and left to solidify at room temperature. Transformants were left for 72 h to regenerate the protoplasts. Transformants were then restreaked according to the procedure described in “strain construction” above.
Miniaturized transformation protocol
To speed up and miniaturize the transformation protocol to make it compatible with a 96-well plate format, it was modified according to the process below. Protoplasts were generated by digestion according to the standard protocol. However, only 50 µL protoplasts were used for the transformation, and 1.75 µL of each RNP complex was added alongside 2.5 µg DNA to these protoplasts and the mixture was incubated on ice for 30 min as in the standard protocol. Then, only 212.5 µL TF3 solution was added to the DNA-RNP-protoplast solution, and the entire mixture was spread on a bottom agar plate following the standard washing, using an L-shaped spreader. No top agar was used in the regeneration of the protoplasts. All steps following the plating of protoplasts were the same as described above in the standard protocol. All results reported in the paper followed the standard transformation protocol unless otherwise indicated.
Generation of pyrG mutants
The two transformation protocols above describe how to transform pyrG strains using a linear DNA fixing template. To generate pyrG mutants in the first place, as we demonstrated with five different A. oryzae strains obtained from NRRL, the standard protocol was changed slightly according to the following modifications. No linear DNA was transformed. Only the two RNP complexes targeting the pyrG gene were added to incubate on ice with the protoplasts. At the final step, strains were plated onto top and bottom agar supplemented with 1 mg/mL 5-FOA 2 g/L uracil, and 5 g/L uridine. Following the regeneration of protoplasts, instead of being restreaked on MMA + Met plates for single colonies and then transferred to MMA + Met slants, strains were restreaked on MMA + Met supplemented with 1 mg/mL 5-FOA and 2 g/L uracil and 5 g/L uridine. Surviving colonies were transferred to slants with the same medium. Between two and three colonies of each strain were analyzed for auxotrophy by plating on CDA, CDA + Uracil + Uridine. One of these strains was subjected to further analysis by amplification of the pyrG gene and flanking regions using primers pyrG-2F and pyrG-2R. The 2640-bp amplicon was purified and subjected to sequencing. The sequences were aligned using Snapgene.
Excision of pyrG marker
To set up the excision to remove the pyrG marker via locus-specific recycling, we followed the protocol described in ref. 96 with minor modifications: spore suspensions were generated by adding 0.5–1 mL sterile water to MMA + Met slants harboring single colonies subjected to colony PCR. Slants were vortexed to resuspend conidia, and then 500 µL of suspension harboring between 10 5 and 10 6 conidia/mL was spread onto PDA + 5-FOA + Uridine + Uracil plates. The conidia were spread using an L-shaped spreader and plates were left to dry for 1–2 h. The plates were then incubated at 30 °C for 5–7 days, at which point healthy, robustly growing colonies appeared on the plates. Conidia from individual colonies that appeared healthy were transferred to PDA + 5-FOA + uridine + uracil slants, whereby they were subject to an additional 4–5 days of growth. These strains had the pyrG marker excised. To verify the marker excision, conidia from slants were subjected to colony PCR (as described in standard transformation protocol), or PCR on extracted genomic DNA. To extract genomic DNA, a small amount of conidia was transferred to 300 µL lysis buffer (2% Triton X-100, 1% SDS, 100 mM NaCl, 1 mM EDTA, 10 mM Tris pH 8) in a bead-beating tube (Lysing Matrix Z, MP Biomedicals, catalog#: 116961050-CF). Bead beating was performed for 1 min. Then, samples were incubated at 65 °C for 30 min, vortexing every 10 min. 300 µL of phenol:chloroform:isoamyl alcohol 25:24:1 reagent was then added (Sigma–Aldrich, #P3803) and tubes were vortexed for 5 min and were then centrifuged at max speed for 10 min to separate the layers. 120 µL of the top aqueous layer was transferred to a new tube, and 210 µL of ice-cold 100% ethanol was added to precipitate the DNA. The DNA pellet was washed twice with 70% and was resuspended in 30 µL sterile water. 1 µL was used as the template for PCR reactions using the PHIRE direct plant PCR kit (ThermoFisher, #F130WH).
Whole-genome sequencing, assembly, annotation, and phylogenetic analysis of diverse A. oryzae strains obtained from NRRL
A. oryzae strains subjected to sequencing were obtained from NRRL (NRRL numbers: #2215, #5592, #32614, #1911, #6574). They were grown in GP-glucose medium (50 mL medium in 250 mL flasks) for 72 h prior to harvesting by vacuum filtration and flash-freezing in liquid nitrogen. gDNA extraction, sample quality assessment, DNA library preparation, sequencing, and bioinformatics analysis were conducted at Azenta Life Sciences. Genomic DNA was extracted using DNeasy Plant Mini Kit following manufacturer’s instructions (Qiagen). Genomic DNA was quantified using the Qubit 2.0 Fluorometer (ThermoFisher Scientific). NEBNext® Ultra™ II DNA Library Prep Kit for Illumina, clustering, and sequencing reagents was used throughout the process following the manufacturer’s recommendations. Briefly, the genomic DNA was fragmented by acoustic shearing with a Covaris S220 instrument. Fragmented DNA was cleaned up and end repaired. Adapters were ligated after adenylation of the 3′ends followed by enrichment by limited cycle PCR. DNA libraries were validated using a High Sensitivity D1000 ScreenTape on the Agilent TapeStation (Agilent Technologies) and were quantified using Qubit 2.0 Fluorometer. The DNA libraries were also quantified by real-time PCR (Applied Biosystems). The sequencing library was clustered onto lanes of an Illumina HiSeq 4000 (or equivalent) flow cell. After clustering, the flow cell was loaded onto the Illumina HiSeq instrument according to the manufacturer’s instructions. The samples were sequenced using a 2 × 150 bp Paired End (PE) configuration. Image analysis and base calling were conducted by the HiSeq Control Software (HCS). Raw sequence data (.bcl files) generated from Illumina HiSeq was converted into FastQ files and de-multiplexed using Illumina bcl2FastQ 2.17 software. One mismatch was allowed for index sequence identification.
Assembly, annotation, and prediction of biosynthetic gene clusters
The reads were filtered with TrimmomaticPE version 0.39 97 with the following parameters: LEADING:30 TRAILING:30 MINLEN:120. The filtered reads were used for de novo assembly using the SPAdes 98 genome assembler v3.13.1-1 with the following parameters --careful --cov-cutoff 100. The resulting assemblies were then processed with AUGUSTUS 99 v3.4.0, to obtain coding sequences and protein predictions. Augustus was executed using a gene model for Aspergillus oryzae to identify start and stop codons, introns, and exons. For the prediction of natural product production repertoire of the strains, the assembled genomes and their gene calling files were used for functional annotation and mining for natural products biosynthetic gene clusters using antiSMASH version 7 100 .
Phylogenetic analysis
The taxonomic affiliation of the A. oryzae strains used in this study (tree in Supplementary Fig. 2 ) was defined using a multilocus phylogenetic tree constructed with the genomes of 59 Aspergillus spp. strains which were obtained from the GenBank database. These genomes were processed with AUGUSTUS 99 v3.4.0, to obtain coding sequences and protein predictions. The predicted proteomes of the Aspergillus dataset. Given the closeness of the strains, a genome from a distantly related taxonomic group ( Trichoderma atroviridae ) was added to the dataset to reduce the number of shared orthologs. The core genome was then calculated using BPGA 101 this analysis led to a set 237 conserved proteins that were sorted, aligned 102 , and trimmed 103 , after this process 167 protein sequences remained. They were then concatenated, and an evolutionary model was calculated for each of the 167 protein partitions. Then a phylogenetic tree was calculated with IQtree2 v2.0.7 104 using maximum likelihood with 10,000 bootstrap replicates. The entire process was executed automatically using a script available at https://github.com/WeMakeMolecules/Core-to-Tree .
Computational identification of candidate neutral, highly transcribed integration sites for protein expression
These sites were identified from a dataset deposited under BioProject accession: “ PRJDB8293 ”. This set of Illumina RNAseq data included 18 libraries which were obtained from A. oryzae RIB40 growing in 50 mL cultures in Czapek–Dox liquid medium supplemented with 1% (w/v) Triton X-100 at 30 °C. This dataset was deposited previously by Wong et al. 105 . The specific datasets that were used are shown in Supplementary Table 13 . The reads were downloaded from the GenBank FTP using fastq-dump v2.113, the reads were then aligned to the A. oryzae reference genome (NCBI RefSeq assembly GCF_000184455.2) using subread package v 2.0.3 106 . To select highly expressed genes, we counted the number of reads that were mapped to each gene in the A. oryzae RIB40 genome using featureCounts v2.0.3 107 . The read counts were calculated independently for each run. As the read counts depend on the depth and processing of each sample, a single cutoff cannot be established. Instead, we used this value to rank genes from most expressed to not expressed using the numbers of reads mapped per library (Supplementary Table 13 ). Then we reasoned that gene that ranked top in all libraries, could be safely considered highly expressed. For selection of neutral, highly transcribed integration sites, we first identified all the intergenic regions in the genome and calculated the average read count of the genes flaking them. Then, we sorted these regions from highest to lowest by their average flanking gene read count. For each of the 18 libraries we selected the top 500 intergenic regions and filtered out those that were not found in all conditions; therefore, we selected intergenic regions that are flanked by constitutive, highly expressed genes. This led to a set of 334 regions that were filtered by length (>4.8 kb). Finally, we selected 10 promising intergenic regions spread across A. oryza e (Fig. 2 , Supplementary Table 4 ).
Computational identification of candidate endogenous bidirectional promoters
For the development of new bidirectional promoters for A. oryzae , we used the annotated genome of strain RIB40 to mine for all the coding sequences whose start codons are in opposite directions. Then we selected the gene pairs that were highly expressed in most conditions using the Wong et al dataset 105 described above. We then identified the promoter region. We used the same dataset or read counts used for the identification of neutral, highly transcribed integration sites.
Identification of core promoters for expression in A. oryzae using the synthetic expression system
To identify candidate core promoters from the A. oryzae genome, we searched for highly expressed genes from publicly available transcriptome data 108 . The gene list of A. oryzae RIB40 grown in CD-glucose in liquid cultures without ER stress was sorted by RPKM to generate a list of the top most highly expressed genes. The top eight most highly expressed genes (unique genes, no duplicates) were selected as candidate strong promoters. Additionally, pdcA , which appeared at #15 in the rank of this list was selected as other studies suggest this is one of the most highly expressed genes in A. oryzae 109 . thiA , which was ranked #20 in this list, was also selected, because it has successfully been used for overexpression in A. oryzae previously 49 . Finally, hhfA , which ranked #55 in the list, was selected, as it was part of the p4-2 bidirectional promoter used in this study. For each of these genes, the genetic DNA 200 bp upstream of the start codon was used as core promoter sequence and ordered as synthetic dsDNA for downstream cloning and transformation. Two additional core promoters that did not come from the transcriptome rank analysis and were not native sequences to A. oryzae were also included. These were An_201205 from A. niger , which was used previously in the development of a Synthetic expression system in A. niger and T. reesei 18 , as well as the core promoter for Afl_ecm33 from gene AFL2G_04718 in Aspergillus flavus . Afl_ecm33 has been successfully used before to express a secondary metabolite in A. oryzae (promoter P4 in this study 48 ). For the An_201205 core promoter, the sequence was identical to the one used previously 18 , but for Afl_ecm33, the 200 bp upstream of the start codon were selected as the core promoter.
Flow cytometry assays of conidia for fluorescence quantification
The overall approach followed the method for promoter evaluation described in ref. 47 but with minor modifications. For all flow cytometry assays, strains were grown on PDA + 5-FOA + UU slants (excised neutral loci strains) or PDA slants (all others) for 5–6 days at 30 °C to allow robust development of conidia. Conidia were harvested by the addition of 1 mL of sterile water, followed by vortexing. 250 µL conidial suspension was then transferred to a 96-well plate (Corning, Falcon Tissue Culture Plate, #353072). Flow cytometry assays were performed on the BD Accuri C6 instrument (BD Biosciences) using the following settings: Run limit = 50,000 events, FSC-H threshold <80,000, agitation = 1 cycle every 1 wells. Raw fluorescence data were converted into MEFL (mean equivalents of Fluorescein, for GFP) or METR (mean equivalents of Texas Red, for mCherry), using a fluorescence beads standard (Spherotech, #RCP-30-5A). At least three biological replicates were run for each sample. FlowJo software (version 10) was used to analyze the data.
Fluorescence microscopy
Strains were grown on either CDA, CDA-Leu, or CDA-dextrin for 4–5 days at 30 °C. Fluorescent protein expression was then imaged in mycelia (edge of the colony) using Leica Microscope DM6B (Leica) and the associated Leica LAS X software (v.5.1.0).
Proteomic comparison of mCherry expression across media and promoters
to compare the expression of mCherry under endogenous promoters and the core promoters in the synthetic expression system, conidia from three different transformants per construct were inoculated at 5 × 10 5 conidia in 50 mL of either CD-dextrin or CD-glucose medium in 250 mL Erlenmeyer flasks. Strains were grown for 96 h at 30 °C, 160 rpm shaking. Biomass was harvested by vacuum filtration over Miracloth and was then lyophilized prior to proteomics.
Proteomics analysis
Protein was extracted and tryptic peptides were prepared by following established proteomic sample preparation protocol 110 . Briefly, cell pellets were resuspended in Qiagen P2 Lysis Buffer (Qiagen, Hilden, Germany, Cat.#19052) to promote cell lysis. Proteins were precipitated with addition of 1 mM NaCl and 4× vol acetone, followed by two additional wash with 80% acetone in water. The recovered protein pellet was homogenized by pipetting mixing with 100 mM Ammonium bicarbonate in 20% Methanol. Protein concentration was determined by the DC protein assay (BioRad, Hercules, CA). Protein reduction was accomplished using 5 mM tris 2-(carboxyethyl)phosphine (TCEP) for 30 min at room temperature, and alkylation was performed with 10 mM iodoacetamide (IAM; final concentration) for 30 min at room temperature in the dark. Overnight digestion with trypsin was accomplished with a 1:50 trypsin:total protein ratio. The resulting peptide samples were analyzed on an Agilent 1290 UHPLC system coupled to a Thermo Scientific Orbitrap Exploris 480 mass spectrometer for discovery proteomics 111 . Briefly, peptide samples were loaded onto an Ascentis® ES-C18 Column (Sigma–Aldrich, St. Louis, MO) and separated with a 10 min LC gradient from 98% solvent A (0.1% FA in H2O) and 2% solvent B (0.1% FA in ACN) to 65% solvent A and 35% solvent B. Eluting peptides were introduced to the mass spectrometer operating in positive-ion mode and were measured in data-independent acquisition (DIA) mode with a duty cycle of 3 survey scans from m/z 380 to m/z 985 and 45 MS2 scans with precursor isolation width of 13.5 m/z to cover the mass range. DIA raw data files were analyzed by an integrated software suite DIA-NN 112 . The database used in the DIA-NN search (library-free mode) is the latest A. oryzae UniProt proteome FASTA sequences plus the protein sequences of the heterologous proteins and common proteomic contaminants. DIA-NN determines mass tolerances automatically based on first-pass analysis of the samples with automated determination of optimal mass accuracies. The retention time extraction window was determined individually for all MS runs analyzed via the automated optimization procedure implemented in DIA-NN. Protein inference was enabled, and the quantification strategy was set to Robust LC = High Accuracy. Output main DIA-NN reports were filtered with a global FDR = 0.01 on both the precursor level and protein group level. The total peak area of tryptic peptides of identified proteins was used to plot the quantity of the targeted proteins in the samples.
Extraction and LC–MS analysis of ergothioneine and heme in fungal mycelium and reference samples
Extraction and analysis of heme.
Extraction was performed according to the protocol specified in ref. 113 , with minor modifications. Lyophilized fungal biomass was ground into a homogeneous powder using a mortar and pestle. Approximately 30 mg of the powder was then transferred to a bead-beating tube (Lysing Matrix Z, MP Biomedicals, catalog#: 116961050-CF) and 1 mL of TE buffer (10 mM Tris, 1 mM EDTA, pH 8) was added. The tube was vortexed to suspend the powder and was then subjected to bead beating for 2 × 1 min using the Biospec Mini Beadbeater. 750 µL of the bead-beaten solution was then transferred to 15 mL conical tubes containing 4 mL of 8:2 acetonitrile:1.7 M HCl. The tubes were then vortexed for 20 min. Then, 1 mL of saturated 0.25 g MgSO 4 •7H 2 O was added to each tube, followed by the addition of 0.1 g NaCl. This created a separation of the aqueous and organic layers. The tubes were then vortexed for 10 min, followed by spinning down at 5000 rcf for 10 min to separate the layers. 100 µL of the top layer was transferred to an LC–MS vial for analysis. In addition to the wild-type and engineered biomass, we extracted heme from lyophilized plant-based ground beef (IMPOSSIBLE Foods Inc, 12 oz IMPOSSIBLE™ burger made from plants).
LC–MS analysis of heme
For the LC–MS analysis, analytes were chromatographically separated with a Kinetex XB-C18 column (50-mm length, 2.1-mm internal diameter, 2.6-µm particle size; Phenomenex, Torrance, CA) at 50 °C using a 1260 Infinity HPLC system (Agilent Technologies). The injection volume for each measurement was 5 µL. The mobile phase was composed of 0.1% formic acid in water (as mobile phase A) and 0.1% formic acid in acetonitrile (as mobile phase B). Analytes were separated via the following gradient: linearly increased from 20%B to 45.5%B in 1.7 min, linearly increased from 45.5%B to 95%B in 0.2 min, held at 95%B for 1.6 min, linearly decreased from 95%B to 20%B in 0.2 min, held at 20%B for 1.3 min. A flow rate of 1 mL/min was used throughout. The total run time was 5 min. The HPLC system was coupled to an Agilent Technologies 6520 Quadrupole Time-of-Flight Mass Spectrometer (QTOF-MS) with a 1:4 post-column split. Nitrogen gas was used as both the nebulizing and drying gas to facilitate the production of gas-phase ions. Drying and nebulizing gases were set to 10 L/min and 30 psi, respectively, and a drying gas temperature of 330 °C was used throughout. Fragmentor, skimmer, and OCT1 RF voltages were set to 250 V, 65 V, and 400 V, respectively. Electrospray ionization (ESI) was conducted in the positive-ion mode with a capillary voltage of 3.5 kV. MS experiments were carried out in the full-scan mode ( m/z 60–1100) at 0.86 spectra per second for the detection of [M + H] + ions. The instrument was tuned for a range of m/z 50–1700. Prior to LC-ESI-TOF MS analysis, the TOF MS was calibrated with the Agilent ESI-Low TOF tuning mix. Mass accuracy was maintained via reference ion mass correction, which was performed with purine and HP-0921 (Agilent Technologies). Data acquisition was carried out by MassHunter Workstation Software version B.08.00 (Agilent Technologies). Data processing was carried out by MassHunter Workstation Qualitative Analysis version B.06.00 and MassHunter Quantitative Analysis version 10.00. External calibration curves were used to quantify the analytes. Hemin chloride (Sigma–Aldrich, #3741) was used as the standard. The mass spectrum in the standards and samples corresponded to that from other experimentally validated studies of intracellular heme 114 . Calculated concentrations obtained from LC–MS analysis were normalized to the initial dry sample weight used for extraction.
Extraction and analysis of ergothioneine
All samples were lyophilized prior to analysis. For extraction from solid, samples were ground into a fine powder using a mortar and pestle and then approximately 30 mg was transferred to tubes for homogenization (Lysing Matrix Z, MP Biomedicals, catalog#: 116961050-CF). 1 mL of 20% methanol with 0.1% formic acid was added and samples were subjected to bead beating for 2 × 1 min. Following bead-beating, samples were spun down at 12,000 RCF for 10 min to separate the solids. 500 µL supernatant was transferred to a centrifugal spin filter to remove any particulates larger molecules (3 kDa cutoff) (Amicon Ultra, Sigma–Aldrich, Catalog # UFC500324). The flow-through was collected and subjected to analysis by LC–MS. In addition to wild-type and engineered fungal biomass, we extracted ergothioneine from the fruiting body of the oyster mushroom ( Pleurotus ostreatus ). The mushroom was purchased fresh (from Berkeley Bowl in Berkeley, CA) and subjected to lyophilization prior to extraction using the procedure above.
LC–MS analysis of ergothioneine
For LC–MS analysis, analytes were chromatographically separated with a Kinetex HILIC column (100-mm length, 4.6-mm internal diameter, 2.6-µm particle size; Phenomenex, Torrance, CA) at 20 °C using a 1260 Infinity HPLC system (Agilent Technologies, Santa Clara, CA, USA). The injection volume for each measurement was 2 µL. The mobile phase was composed of 10 mM ammonium formate (prepared from a pre-made solution from Sigma–Aldrich, St. Louis, MO, USA) and 0.2% formic acid (from an original stock at ≥98% chemical purity from Sigma–Aldrich) in water (as mobile phase A) and 10 mM ammonium formate and 0.2% formic acid in 90% acetonitrile with the remaining solvent being water (as mobile phase B). The solvents used were of LC–MS grade and purchased from Honeywell Burdick & Jackson, CA, USA. Analytes were separated via the following gradient: linearly decreased from 90%B to 70%B in 4 min, held at 70%B for 1.5 min, linearly decreased from 70%B to 40%B in 0.5 min, held at 40%B for 2.5 min, linearly increased from 40%B to 90%B in 0.5 min, held at 90%B for 2 min. The flow rate was changed as follows: 0.6 mL/min for 6.5 min, linearly increased from 0.6 mL/min to 1 mL/min for 0.5 min, held at 1 mL/min for 4 min. The total run time was 11 min. The HPLC system was coupled to an Agilent Technologies 6520 Quadrupole Time-of-Flight Mass Spectrometer (QTOF-MS) with a 1:4 post-column split. Nitrogen gas was used as both the nebulizing and drying gas to facilitate the production of gas-phase ions. Drying and nebulizing gases were set to 12 L/min and 25 psi, respectively, and a drying gas temperature of 350 °C was used throughout. Fragmentor, skimmer, and OCT1 RF voltages were set to 100 V, 50 V, and 250 V, respectively. Electrospray ionization (ESI) was conducted in the positive-ion mode with a capillary voltage of 3.5 kV. MS experiments were carried out in the full-scan mode ( m/z 70–1100) at 0.86 spectra per second for the detection of [M + H] + ions. The instrument was tuned for a range of m/z 50–1700. Prior to LC-ESI-TOF MS analysis, the TOF MS was calibrated with the Agilent ESI-Low TOF tuning mix. Mass accuracy was maintained via reference ion mass correction, which was performed with purine and HP-0921 (Agilent Technologies). Data acquisition was carried out by MassHunter Workstation Software version B.08.00 (Agilent Technologies). Data processing was carried out by MassHunter Workstation Qualitative Analysis version B.06.00 and MassHunter Quantitative Analysis version 10.00. External calibration curves were used to quantify the analytes. Ergothioneine (Sigma–Aldrich, #7521-25MG) was used as the standard. The mass spectrum in the standards and samples corresponded to that from other experimentally validated studies of intracellular ergothioneine 115 . Calculated concentrations obtained from LC–MS analysis were normalized to the initial dry sample weight used for extraction.
Protein and amino acid analysis in fungal mycelium
Protein content was analyzed by combustion method, directly following the Method 990.03 described in ref. 116 . The combustion was performed using Leco FP-528 Nitrogen Combustion Analyzer (Leco). Crude Protein was calculated as Nitrogen × 6.25. Amino acid composition was analyzed using acid hydrolysis of lyophilized fungal biomass, directly following the protocols of Method 994.12 described in ref. 116 , Method 982.30 in ref. 117 , as well as the methods described in ref. 118 .
Statistics and Reproducibility
No statistical method was used to predetermine sample size. n = 3 was chosen as the minimal number of replicates for experimental characterization. We determined this to be sufficient based on internal controls (using previously characterized promoters and fluorescent genes) to capture biological variability between transformants/strains. All microscopy images and PCR results for confirming insertion/excision displayed are representative of at least three biological replicates. No data were excluded from the analyses. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper, supplementary information, the supplementary data files, and in the source data file. Source data are provided as a Source Data file. Strains and plasmids (and their associated sequences) generated in this study have been deposited in the JBEI Public Registry ( https://public-registry.jbei.org/ ). See Supplementary Tables 11 – 12 for plasmids and strain information. Outputs of computational analysis for identification of candidate endogenous neutral loci and bidirectional promoters are available in Supplementary data files 1 and 2. Output of mass spectrometry data are available as source data. The generated mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE 94 partner repository with the dataset identifier “ PXD043152 ”. The genome sequences of the A. oryzae strains obtained from NRRL and sequenced as part of this study have been deposited to the Sequence Read Archive under Bioproject “ PRJNA987873 ”. The previously published transcriptome data used to identify endogenous neutral loci and bidirectional promoters in Aspergillus oryzae can be found in GenBank under BioProject accession: “ PRJDB8293 ”. CAoGD (Comprehensive Aspergillus oryzae Genome Database) v.2.4 was used to identify the genomic location and sequence identity of endogenous promoters and genes targeted in transformations ( https://nribf21.nrib.go.jp/CAoGD/ ). Source data are provided with this paper.
Code availability
No custom code was used in the analysis of data, and all the previously published software used as well as the relevant commands has been cited in the methods. The automatic phylogenomic analysis from the core genomes of Aspergillus oryzae strains was executed using the script available at https://github.com/WeMakeMolecules/Core-to-Tree . DIA-NN is freely available for download from https://github.com/vdemichev/DiaNN .
Crippa, M. et al. Food systems are responsible for a third of global anthropogenic GHG emissions. Nat. Food 2 , 198–209 (2021).
Article CAS PubMed Google Scholar
Springmann, M. et al. Options for keeping the food system within environmental limits. Nature 562 , 519–525 (2018).
Article ADS CAS PubMed Google Scholar
Waage, J. et al. Changing food systems and infectious disease risks in low-income and middle-income countries. Lancet Planet. Health 6 , e760–e768 (2022).
Article PubMed PubMed Central Google Scholar
Graham, A. E. & Ledesma-Amaro, R. The microbial food revolution. Nat. Commun. 14 , 2231 (2023).
Article ADS CAS PubMed PubMed Central Google Scholar
Jahn, L. J., Rekdal, V. M. & Sommer, M. O. A. Microbial foods for improving human and planetary health. Cell 186 , 469–478 (2023).
Humpenoder, F. et al. Projected environmental benefits of replacing beef with microbial protein. Nature 605 , 90–96 (2022).
Article ADS PubMed Google Scholar
van Dijk, M., Morley, T., Rau, M. L. & Saghai, Y. A meta-analysis of projected global food demand and population at risk of hunger for the period 2010–2050. Nat. Food 2 , 494–501 (2021).
Article PubMed Google Scholar
Javourez, U., Rosero Delgado, E. A. & Hamelin, L. Upgrading agrifood co-products via solid fermentation yields environmental benefits under specific conditions only. Nat. Food 3 , 911–920 (2022).
Jarvio, N. et al. Ovalbumin production using Trichoderma reesei culture and low-carbon energy could mitigate the environmental impacts of chicken-egg-derived ovalbumin. Nat. Food 2 , 1005–1013 (2021).
Tang, X. et al. Characterization of an omega-3 desaturase from Phytophthora parasitica and application for eicosapentaenoic acid production in Mortierella alpina. Front. Microbiol. 9 , 1878 (2018).
Wiebe, M. G. Myco-protein from Fusarium venenatum: a well-established product for human consumption. Appl. Microbiol. Biotechnol. 58 , 421–427 (2002).
Meyer, V. et al. Growing a circular economy with fungal biotechnology: a white paper. Fungal Biol. Biotechnol. 7 , 5 (2020).
Yamashita, H. Koji starter and koji world in Japan. J. Fungi. https://doi.org/10.3390/jof7070569 (2021).
Aro, N. et al. Production of bovine beta-lactoglobulin and hen egg ovalbumin by Trichoderma reesei using precision fermentation technology and testing of their techno-functional properties. Food Res. Int. 163 , 112131 (2023).
Wu, V. W. et al. The regulatory and transcriptional landscape associated with carbon utilization in a filamentous fungus. Proc. Natl Acad. Sci. USA 117 , 6003–6013 (2020).
Huttner, S., Johansson, A., Goncalves Teixeira, P., Achterberg, P. & Nair, R. B. Recent advances in the intellectual property landscape of filamentous fungi. Fungal Biol. Biotechnol. 7 , 16 (2020).
Denby, C. M. et al. Industrial brewing yeast engineered for the production of primary flavor determinants in hopped beer. Nat. Commun. 9 , 965 (2018).
Article ADS PubMed PubMed Central Google Scholar
Rantasalo, A. et al. A universal gene expression system for fungi. Nucleic Acids Res. 46 , e111 (2018).
Gamarra-Castillo, O., Echeverry-Montana, N., Marbello-Santrich, A., Hernandez-Carrion, M. & Restrepo, S. Meat substitute development from fungal protein (Aspergillus oryzae). Foods https://doi.org/10.3390/foods11192940 (2022).
Prime Roots, I. Prime Roots Website https://www.primeroots.com/ (2023).
Liu, L., Feizi, A., Osterlund, T., Hjort, C. & Nielsen, J. Genome-scale analysis of the high-efficient protein secretion system of Aspergillus oryzae. BMC Syst. Biol. 8 , 73 (2014).
Merz, M. et al. Flavourzyme, an enzyme preparation with industrial relevance: automated nine-step purification and partial characterization of eight enzymes. J. Agric. Food Chem. 63 , 5682–5693 (2015).
Ogawa, M., Moreno Garcia, J., Nitin, N., Baar, K. & Block, D. E. Assessing edible filamentous fungal carriers as cell supports for growth of yeast and cultivated meat. Foods https://doi.org/10.3390/foods11193142 (2022).
Katayama, T. et al. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnol. Lett. 38 , 637–642 (2016).
Yoon, J., Maruyama, J. & Kitamoto, K. Disruption of ten protease genes in the filamentous fungus Aspergillus oryzae highly improves production of heterologous proteins. Appl. Microbiol. Biotechnol. 89 , 747–759 (2011).
Zhang, J. et al. Ku80 gene is related to non-homologous end-joining and genome stability in Aspergillus niger. Curr. Microbiol. 62 , 1342–1346 (2011).
Leynaud-Kieffer, L. M. C. et al. A new approach to Cas9-based genome editing in Aspergillus niger that is precise, efficient and selectable. PLoS ONE 14 , e0210243 (2019).
Article CAS PubMed PubMed Central Google Scholar
Katayama, T. et al. Forced recycling of an AMA1-based genome-editing plasmid allows for efficient multiple gene deletion/integration in the industrial filamentous fungus Aspergillus oryzae. Appl. Environ. Microbiol. https://doi.org/10.1128/AEM.01896-18 (2019).
Jan Vonk, P., Escobar, N., Wosten, H. A. B., Lugones, L. G. & Ohm, R. A. High-throughput targeted gene deletion in the model mushroom Schizophyllum commune using pre-assembled Cas9 ribonucleoproteins. Sci. Rep. 9 , 7632 (2019).
Article ADS Google Scholar
Tong, S. et al. Evasion of Cas9 toxicity to develop an efficient genome editing system and its application to increase ethanol yield in Fusarium venenatum TB01. Appl. Microbiol. Biotechnol. 106 , 6583–6593 (2022).
Schuster, M. & Kahmann, R. CRISPR-Cas9 genome editing approaches in filamentous fungi and oomycetes. Fungal Genet. Biol. 130 , 43–53 (2019).
Foster, A. J. et al. CRISPR-Cas9 ribonucleoprotein-mediated co-editing and counterselection in the rice blast fungus. Sci. Rep. 8 , 14355 (2018).
Abdallah, Q. A., Ge, W. & Fortwendel, J. R. A simple and universal system for gene manipulation in Aspergillus fumigatus: in vitro-assembled Cas9-guide RNA ribonucleoproteins coupled with microhomology repair templates. mSphere https://doi.org/10.1128/msphere.00446-00417 (2017).
Pohl, C., Kiel, J. A. K. W., Driessen, A. J. M., Bovenberg, R. A. L. & Nygård, Y. CRISPR/Cas9 based genome editing of Penicillium chrysogenum. ACS Synth. Biol. 5 , 754–764 (2016).
Kuivanen, J., Korja, V., Holmström, S. & Richard, P. Development of microtiter plate scale CRISPR/Cas9 transformation method for Aspergillus niger based on in vitro assembled ribonucleoprotein complexes. Fungal Biol. Biotechnol. 6 , 3 (2019).
Hao, Z. & Su, X. Fast gene disruption in Trichoderma reesei using in vitro assembled Cas9/gRNA complex. BMC Biotechnol. 19 , 2 (2019).
Maruyama, J. & Kitamoto, K. Multiple gene disruptions by marker recycling with highly efficient gene-targeting background (DeltaligD) in Aspergillus oryzae. Biotechnol. Lett. 30 , 1811–1817 (2008).
Zhang, C., Meng, X., Wei, X. & Lu, L. Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal Genet. Biol. 86 , 47–57 (2016).
Rantasalo, A. et al. Novel genetic tools that enable highly pure protein production in Trichoderma reesei. Sci. Rep. 9 , 5032 (2019).
Liu, K. et al. Dual sgRNA-directed gene deletion in basidiomycete Ganoderma lucidum using the CRISPR/Cas9 system. Microb. Biotechnol. 13 , 386–396 (2020).
Gao, D., Smith, S., Spagnuolo, M., Rodriguez, G. & Blenner, M. Dual CRISPR-Cas9 cleavage mediated gene excision and targeted integration in Yarrowia lipolytica. Biotechnol. J. 13 , 1700590 (2018).
Article Google Scholar
Han, H. et al. High-efficient production of mushroom polyketide compounds in a platform host Aspergillus oryzae. Micro. Cell Fact. 22 , 60 (2023).
Article CAS Google Scholar
Weld, R. J., Plummer, K. M., Carpenter, M. A. & Ridgway, H. J. Approaches to functional genomics in filamentous fungi. Cell Res. 16 , 31–44 (2006).
Chaves, J. E. et al. Evaluation of chromosomal insertion loci in the Pseudomonas putida KT2440 genome for predictable biosystems design. Metab. Eng. Commun. 11 , e00139 (2020).
Reider Apel, A. et al. A Cas9-based toolkit to program gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 45 , 496–508 (2017).
Liu, C. et al. Efficient reconstitution of Basidiomycota diterpene erinacine gene cluster in Ascomycota host Aspergillus oryzae based on genomic DNA sequences. J. Am. Chem. Soc. 141 , 15519–15523 (2019).
Wei, P. L. et al. Quantitative characterization of filamentous fungal promoters on a single-cell resolution to discover cryptic natural products. Sci. China Life Sci. 66 , 848–860 (2023).
Umemura, M., Kuriiwa, K., Dao, L. V., Okuda, T. & Terai, G. Promoter tools for further development of Aspergillus oryzae as a platform for fungal secondary metabolite production. Fungal Biol. Biotechnol. 7 , 3 (2020).
Shoji, J. Y., Maruyama, J., Arioka, M. & Kitamoto, K. Development of Aspergillus oryzae thiA promoter as a tool for molecular biological studies. FEMS Microbiol. Lett. 244 , 41–46 (2005).
Kanemori, Y., Gomi, K., Kitamoto, K., Kumagai, C. & Tamura, G. Insertion analysis of putative functional elements in the promoter region of the Aspergillus oryzae Taka-amylase A gene (amyB) using a heterologous Aspergillus nidulans amdS-lacZ fusion gene system. Biosci. Biotechnol. Biochem. 63 , 180–183 (1999).
Liu, Q. et al. A programmable high-expression yeast platform responsive to user-defined signals. Sci. Adv. 8 , eabl5166 (2022).
Sheets, M. B., Tague, N. & Dunlop, M. J. An optogenetic toolkit for light-inducible antibiotic resistance. Nat. Commun. 14 , 1034 (2023).
Brophy, J. A. N. et al. Synthetic genetic circuits as a means of reprogramming plant roots. Science 377 , 747–751 (2022).
Sanford, A., Kiriakov, S. & Khalil, A. S. A toolkit for precise, multigene control in Saccharomyces cerevisiae. ACS Synth. Biol. 11 , 3912–3920 (2022).
Li, H. S. et al. Multidimensional control of therapeutic human cell function with synthetic gene circuits. Science 378 , 1227–1234 (2022).
Mozsik, L. et al. Modular synthetic biology toolkit for Filamentous Fungi. ACS Synth. Biol. 10 , 2850–2861 (2021).
Belcher, M. S. et al. Design of orthogonal regulatory systems for modulating gene expression in plants. Nat. Chem. Biol. 16 , 857–865 (2020).
Wanka, F. et al. Tet-on, or Tet-off, that is the question: advanced conditional gene expression in Aspergillus. Fungal Genet. Biol. 89 , 72–83 (2016).
Helmschrott, C., Sasse, A., Samantaray, S., Krappmann, S. & Wagener, J. Upgrading fungal gene expression on demand: improved systems for doxycycline-dependent silencing in Aspergillus fumigatus. Appl. Environ. Microbiol. 79 , 1751–1754 (2013).
Gomi, K. Regulatory mechanisms for amylolytic gene expression in the koji mold Aspergillus oryzae. Biosci. Biotechnol. Biochem. 83 , 1385–1401 (2019).
Elison, G. L., Xue, Y., Song, R. & Acar, M. Insights into bidirectional gene expression control using the canonical GAL1/GAL10 promoter. Cell Rep. 25 , 737–748.e734 (2018).
Bowman, E. K. et al. Bidirectional titration of yeast gene expression using a pooled CRISPR guide RNA approach. Proc. Natl Acad. Sci. USA 117 , 18424–18430 (2020).
Rantasalo, A. et al. Synthetic transcription amplifier system for orthogonal control of gene expression in Saccharomyces cerevisiae. PLoS ONE 11 , e0148320 (2016).
Rendsvig, J. K. H., Workman, C. T. & Hoof, J. B. Bidirectional histone-gene promoters in Aspergillus: characterization and application for multi-gene expression. Fungal Biol. Biotechnol. 6 , 24 (2019).
Halliwell, B. & Cheah, I. Ergothioneine, where are we now? FEBS Lett. 596 , 1227–1230 (2022).
Beelman, R. B. et al. Health consequences of improving the content of ergothioneine in the food supply. FEBS Lett. 596 , 1231–1240 (2022).
Bello, M. H., Barrera-Perez, V., Morin, D. & Epstein, L. The Neurospora crassa mutant NcDeltaEgt-1 identifies an ergothioneine biosynthetic gene and demonstrates that ergothioneine enhances conidial survival and protects against peroxide toxicity during conidial germination. Fungal Genet. Biol. 49 , 160–172 (2012).
Irani, S. et al. Snapshots of C-S cleavage in Egt2 reveals substrate specificity and reaction mechanism. Cell Chem. Biol. 25 , 519–529.e514 (2018).
Hu, W. et al. Bioinformatic and biochemical characterizations of C-S bond formation and cleavage enzymes in the fungus Neurospora crassa ergothioneine biosynthetic pathway. Org. Lett. 16 , 5382–5385 (2014).
Takusagawa, S., Satoh, Y., Ohtsu, I. & Dairi, T. Ergothioneine production with Aspergillus oryzae. Biosci. Biotechnol. Biochem. 83 , 181–184 (2019).
Devaere, J. et al. Improving the aromatic profile of plant-based meat alternatives: effect of myoglobin addition on volatiles. Foods https://doi.org/10.3390/foods11131985 (2022).
Fraser, R. Z., Shitut, M., Agrawal, P., Mendes, O. & Klapholz, S. Safety evaluation of soy leghemoglobin protein preparation derived from Pichia pastoris, intended for use as a flavor catalyst in plant-based meat. Int. J. Toxicol. 37 , 241–262 (2018).
Voigt, C. A. Synthetic biology 2020-2030: six commercially-available products that are changing our world. Nat. Commun. 11 , 6379 (2020).
Food and Drug Administration. GRAS Notice no. GRN 001001: Myoglobin preparation from Bos taurus https://www.fda.gov/media/153921/download (2021).
Franken, A. C. et al. Heme biosynthesis and its regulation: towards understanding and improvement of heme biosynthesis in filamentous fungi. Appl. Microbiol. Biotechnol. 91 , 447–460 (2011).
Ishchuk, O. P. et al. Genome-scale modeling drives 70-fold improvement of intracellular heme production in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 119 , e2108245119 (2022).
Michener, J. K., Nielsen, J. & Smolke, C. D. Identification and treatment of heme depletion attributed to overexpression of a lineage of evolved P450 monooxygenases. Proc. Natl Acad. Sci. USA 109 , 19504–19509 (2012).
Liu, L., Martinez, J. L., Liu, Z., Petranovic, D. & Nielsen, J. Balanced globin protein expression and heme biosynthesis improve production of human hemoglobin in Saccharomyces cerevisiae. Metab. Eng. 21 , 9–16 (2014).
Elrod, S. L., Jones, A., Berka, R. M. & Cherry, J. R. Cloning of the Aspergillus oryzae 5-aminolevulinate synthase gene and its use as a selectable marker. Curr. Genet. 38 , 291–298 (2000).
Franken, A. C. et al. Analysis of the role of the Aspergillus niger aminolevulinic acid synthase (hemA) gene illustrates the difference between regulation of yeast and fungal haem- and sirohaem-dependent pathways. FEMS Microbiol. Lett. 335 , 104–112 (2012).
Zhou, S. et al. Heme-biosynthetic porphobilinogen deaminase protects Aspergillus nidulans from nitrosative stress. Appl. Environ. Microbiol. 78 , 103–109 (2012).
Gonzalez-Dominguez, M., Freire-Picos, M. A. & Cerdan, M. E. Haem regulation of the mitochondrial import of the Kluyveromyces lactis 5-aminolaevulinate synthase: an organelle approach. Yeast 18 , 41–48 (2001).
Zoładek, T., Nguyen, B. N. & Rytka, J. Saccharomyces cerevisiae mutants defective in heme biosynthesis as a tool for studying the mechanism of phototoxicity of porphyrins. Photochem. Photobio. 64 , 957–962 (1996).
Twala, P. P., Mitema, A., Baburam, C. & Feto, N. A. Breakthroughs in the discovery and use of different peroxidase isoforms of microbial origin. AIMS Microbiol. 6 , 330–349 (2020).
Zhao, F. et al. Multiplex base-editing enables combinatorial epigenetic regulation for genome mining of fungal natural products. J. Am. Chem. Soc. 145 , 413–421 (2023).
Jarczynska, Z. D. et al. A versatile in vivo DNA assembly toolbox for fungal strain engineering. ACS Synth. Biol. 11 , 3251–3263 (2022).
Jarczynska, Z. D. et al. DIVERSIFY: a fungal multispecies gene expression platform. ACS Synth. Biol. 10 , 579–588 (2021).
van der Hoek, S. A. et al. Engineering ergothioneine production in Yarrowia lipolytica. FEBS Lett. 596 , 1356–1364 (2022).
van der Hoek, S. A. et al. Engineering precursor supply for the high-level production of ergothioneine in Saccharomyces cerevisiae. Metab. Eng. 70 , 129–142 (2022).
Watarai, N., Yamamoto, N., Sawada, K. & Yamada, T. Evolution of Aspergillus oryzae before and after domestication inferred by large-scale comparative genomic analysis. DNA Res. 26 , 465–472 (2019).
Ropars, J. et al. Domestication of the emblematic white cheese-making fungus Penicillium camemberti and its diversification into two varieties. Curr. Biol. 30 , 4441–4453.e4444 (2020).
Wu, F. et al. Opinion: allow golden rice to save lives. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.2120901118 (2021).
Waltz, E. GABA-enriched tomato is first CRISPR-edited food to enter market. Nat. Biotechnol. 40 , 9–11 (2022).
Ham, T. S. et al. Design, implementation and practice of JBEI-ICE: an open source biological part registry platform and tools. Nucleic Acids Res. 40 , e141 (2012).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6 , 343–345 (2009).
Nemoto, T., Maruyama, J. & Kitamoto, K. Contribution ratios of amyA, amyB, amyC genes to high-level alpha-amylase expression in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 76 , 1477–1483 (2012).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 , 2114–2120 (2014).
Prjibelski, A., Antipov, D., Meleshko, D., Lapidus, A. & Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 70 , e102 (2020).
Stanke, M., Diekhans, M., Baertsch, R. & Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24 , 637–644 (2008).
Blin, K. et al. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 51 , W46–W50 (2023).
Chaudhari, N. M., Gupta, V. K. & Dutta, C. BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6 , 24373 (2016).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 , 1792–1797 (2004).
Talavera, G. & Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56 , 564–577 (2007).
Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for Phylogenetic inference in the genomic era. Mol. Biol. Evol. 37 , 1530–1534 (2020).
Wong, P. S., Tamano, K. & Aburatani, S. Improvement of free fatty acid secretory productivity in Aspergillus oryzae by comprehensive analysis on time-series gene expression. Front. Microbiol. 12 , 605095 (2021).
Liao, Y., Smyth, G. K. & Shi, W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41 , e108 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30 , 923–930 (2013).
Wang, B. et al. Survey of the transcriptome of Aspergillus oryzae via massively parallel mRNA sequencing. Nucleic Acids Res. 38 , 5075–5087 (2010).
Liu, P., Lim, J. Y., Kim, H. S., Kim, J. H. & Chae, K. S. Isolation and characterization of the mheA (most highly expressed) gene of Aspergillus oryzae. Mycobiology 40 , 208–209 (2012).
Chen, Y., Gin, J. & Petzold, J. C. Alkaline-SDS cell lysis of microbes with acetone protein precipitation for proteomic sample preparation in 96-well plate format. dx. https://doi.org/10.17504/protocols.io.6qpvr6xjpvmk/v1 (2023).
Chen, Y., Gin, J. & Petzold, J. C. Discovery proteomic (DIA) LC-MS/MS data acquisition and analysis. dx. https://doi.org/10.17504/protocols.io.6qpvr6xjpvmk/v1 (2022).
Demichev, V., Messner, C. B., Vernardis, S. I., Lilley, K. S. & Ralser, M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 17 , 41–44 (2020).
Fyrestam, J. & Ostman, C. Determination of heme in microorganisms using HPLC-MS/MS and cobalt(III) protoporphyrin IX inhibition of heme acquisition in Escherichia coli. Anal. Bioanal. Chem. 409 , 6999–7010 (2017).
Sana, T. R., Waddell, K. & Fischer, S. M. A sample extraction and chromatographic strategy for increasing LC/MS detection coverage of the erythrocyte metabolome. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 871 , 314–321 (2008).
Wang, L. Z. et al. Quantification of L-ergothioneine in human plasma and erythrocytes by liquid chromatography-tandem mass spectrometry. J. Mass Spectrom. 48 , 406–412 (2013).
The Association of Official Analytical Chemists. Official Methods of Analysis 17th edn (AOAC International, Gaithersburg, Md, 2000). https://search.worldcat.org/title/official-methods-of-analysis-of-aoac-international/oclc/476032693 .
The Association of Official Analytical Chemists. Official Methods of Analysis 18th edn (AOAC International, Gaithersburg, Md, 2006). https://search.worldcat.org/title/officialmethods-of-analysis-of-aoac-international/oclc/62751475 .
Gehrke, C. W., Rexroad, P. R., Schisla, R. M., Absheer, J. S. & Zumwalt, R. W. Quantitative analysis of cystine, methionine, lysine, and nine other amino acids by a single oxidation-4 hour hydrolysis method. J. Assoc. Anal. Chem. 70 , 171–174 (1987).
CAS Google Scholar
Download references
Acknowledgements
V.M.R. was supported by the Miller Institute at the University of California, Berkeley. P.C.M. and J.D.K. were supported by Novo Nordisk Foundation grant no. NNF20CC0035580. C.V.D.L. was supported by Novo Nordisk Foundation grant NNF21OC0065495. This work was part of the DOE Joint BioEnergy Institute ( https://www.jbei.org ) supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy.
Author information
Authors and affiliations.
Department of Bioengineering, University of California, Berkeley, CA, 94720, USA
Vayu Maini Rekdal & Jay D. Keasling
Miller Institute for Basic Research in Science, University of California, Berkeley, CA, 94720, USA
Vayu Maini Rekdal
Joint BioEnergy Institute, Emeryville, CA, 94608, USA
Vayu Maini Rekdal, Casper R. B. van der Luijt, Yan Chen, Ramu Kakumanu, Edward E. K. Baidoo, Christopher J. Petzold & Jay D. Keasling
Novo Nordisk Foundation Center for Biosustainability, Technical University of Denmark, 2800, Kgs. Lyngby, Denmark
Casper R. B. van der Luijt, Pablo Cruz-Morales & Jay D. Keasling
Department of Food Science, University of Copenhagen, 1958, Frederiksberg, Denmark
Casper R. B. van der Luijt
Lawrence Berkeley National Laboratory, Biological Systems and Engineering Division, Berkeley, CA, 94720, USA
Casper R. B. van der Luijt, Yan Chen, Ramu Kakumanu, Edward E. K. Baidoo, Christopher J. Petzold & Jay D. Keasling
California Institute of Quantitative Biosciences (QB3), University of California, Berkeley, CA, 94720, USA
Jay D. Keasling
Department of Chemical and Biomolecular Engineering, University of California, Berkeley, CA, 94720, USA
You can also search for this author in PubMed Google Scholar
Contributions
VMR and JDK conceptualized the study. VMR developed and optimized transformation methods, established the neutral loci, endogenous promoters, and vectors for transformation, and engineered the final strains. CVDL evaluated parts for the Synthetic expression system and conducted transformation of A. oryzae NRRL strains. PCM conducted bioinformatics analysis, including genome annotation and assembly, phylogenetic analysis, and identification of endogenous neutral loci and bidirectional promoters. YC and CJP conducted proteomics experiments and analysis. RK and EEKB conducted targeted LC–MS experiments and analysis. All authors approved the final manuscript.
Corresponding author
Correspondence to Jay D. Keasling .
Ethics declarations
Competing interests.
J.D.K. has financial interests in Amyris, Ansa Biotechnologies, Apertor Pharma, Berkeley Yeast, Cyklos Materials, Demetrix, Lygos, Napigen, ResVita Bio, and Zero Acre Farms. V.M.R. and J.D.K. are listed as inventors on a provisional patent (US22/42816) which relates to the methods composition described in the engineering of heme metabolism in edible fungal biomass. The other authors declare no competing interests.
Peer review
Peer review information.
Nature Communications thanks Josephine Wee, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information, peer review file, description of additional supplementary files, supplementary data 1, supplementary data 2, reporting summary, source data, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Maini Rekdal, V., van der Luijt, C.R.B., Chen, Y. et al. Edible mycelium bioengineered for enhanced nutritional value and sensory appeal using a modular synthetic biology toolkit. Nat Commun 15 , 2099 (2024). https://doi.org/10.1038/s41467-024-46314-8
Download citation
Received : 29 June 2023
Accepted : 21 February 2024
Published : 14 March 2024
DOI : https://doi.org/10.1038/s41467-024-46314-8
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Translational Research newsletter — top stories in biotechnology, drug discovery and pharma.

IMAGES
VIDEO
COMMENTS
Stem cells are thought to alleviate the damage caused by chemotherapy drugs and to play role... YuSheng Zhang, YaNan Liu, Zi Teng, ZeLin Wang, Peng Zhu, ZhiXin Wang, FuJun Liu and XueXia Liu. Biological Research 2023 56 :47. Research article Published on: 13 August 2023.
Read the latest Research articles in Biological sciences from Scientific Reports. ... Chemical biology; ... Calls for Papers Guide to referees ...
The concept includes anatomy, physiology, cell biology, biochemistry and biophysics, and covers all organisms from microorganisms, animals to plants. ... Latest Research and Reviews.
Syncing brains with your leader. In-group social bonding stabilizes hierarchical structures in various species. Jun Ni, Jiaxin Yang and Yina Ma show that social bonding in humans selectively increases information exchange and prefrontal neural synchronization for human leader-follower pairs. Image credit: pbio.3002545. 03/14/2024.
We are pleased to share with you the 50 most read Nature Communications articles* in life and biological sciences published in 2019. Featuring authors from around the world, these papers highlight ...
Feature papers represent the most advanced research with significant potential for high impact in the field. A Feature Paper should be a substantial original Article that involves several techniques or approaches, provides an outlook for future research directions and describes possible research applications. ... Biology is an international ...
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology.The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and ...
Anne Rosenwald. Explore the latest full-text research PDFs, articles, conference papers, preprints and more on BIOLOGICAL SCIENCE. Find methods information, sources, references or conduct a ...
Current Biology is a general journal that publishes original research across all areas of biology together with an extensive and varied set of editorial sections. A primary aim of the journal is to foster communication across fields of biology, both by publishing important findings of general interest from diverse fields and through highly accessible editorial articles that explicitly aim to ...
The Biology of Cancer. Cancer is a disease that begins with genetic and epigenetic alterations occurring in specific cells, some of which can spread and migrate to other tissues. 4 Although the biological processes affected in carcinogenesis and the evolution of neoplasms are many and widely different, we will focus on 4 aspects that are particularly relevant in tumor biology: genomic and ...
M. Pitkanen. Jan 2024. Davron Maxammadiyev. Nur Hidayah. Explore the latest full-text research PDFs, articles, conference papers, preprints and more on BIOLOGY. Find methods information, sources ...
Cell biology is the discipline of biological sciences that studies the structure, physiology, growth, reproduction and death of cells. Research in cell biology uses microscopic and molecular tools ...
Autophagy, unfolded protein response and lung disease. Mohammad S. Akhter, ... Nektarios Barabutis. 2020 View PDF. More opportunities to publish your research: Read the latest articles of Current Research in Cell Biology at ScienceDirect.com, Elsevier's leading platform of peer-reviewed scholarly literature.
There is no precise definition of artificial intelligence (AI) so far, but in general it refers to the ability of any machines which can simulate the intelligences of higher organisms. The field of AI has important roots in almost every branch of research including philosophy, mathematics, computing, psychology and biology [ 1 ].
Scientific research report format is based on the scientific method and is organized to enable the reader to quickly comprehend the main points of the investigation. The format required in all biology classes consists of a Title, Abstract, Introduction, Methods, Results, Discussion, and Literature Cited sections.
The Journal of heredity. Rushworth, CA; Mitchell-Olds, T. Despite decades of research, the evolution of sex remains an enigma in evolutionary biology. Typically, research addresses the costs of sex and asexuality to characterize the circumstances favoring one reproductive mode. Surprisingly few studies address the influence of common traits ...
17. Statistics , Environmental Education , Science Communication , Conservation Biology. Antioxidant and Medicinal Properties of Mulberry (Morus SP.): A Review. Mulberry is exclusively used for rearing silkworm due to the presence of unique chemo-factors like morin, β-sitosterol in leaves.
The Journal of Mathematical Biology (JOMB) utilizes diverse mathematical disciplines to advance biological understanding. It publishes papers providing new insights through rigorous mathematical analysis or innovative mathematical tools, with a focus on accessibility to biologists. Covers cell biology, genetics, ecology, and more.
Dimitrije Ivančić. Marc Güell. Article Open Access 3 Dec 2021 Nature Communications. Image credit: KTSDESIGN/SCIENCE PHOTO LIBRARY/Getty. Nature Communications ( Nat Commun) ISSN 2041-1723 ...
Biological Psychology Research Paper Topics. Neurobiological mechanisms underlying memory formation. Impact of sleep disorders on cognitive function and mental health. Biological basis of personality traits and behavior. Neural correlates of emotions and emotional disorders. Role of neuroplasticity in brain recovery after injury.
The 100 most downloaded cell and molecular biology research articles published in Scientific Reports in 2022. ... these papers showcase valuable research from an international community.
The topics discussed in the interview cover the development and activities of the Konrad Lorenz Institute for Evolution and Cognition Research as one of the most important theoretical biology centers in the world, the reasons for its inspiring atmosphere, as well as the development of the interests and research work of its longtime president Gerd B. Müller. An important part of this is the ...
SPOTLIGHT ON RESEARCH PAPER OF THE MONTH FEB 2024 The Committee for Graduate Studies in the Faculty of Biology selects the leading scientific article each month from all the scientific articles published for that month. We are pleased to announce that the winner of February 2024 article is Guy Levin from Prof. Gadi Schuster's lab.
A recyclable CRISPR-Cas9 method for efficient gene integration and expression. We selected Aspergillus oryzae (koji mold) as our model edible fungus and engineering target, as this fungus has a ...