- Make an Appointment
- Study Connect
- Request Workshop


How to read and understand a scientific paper
How to read and understand a scientific paper: a guide for non-scientists, london school of economics and political science, jennifer raff.
From vaccinations to climate change, getting science wrong has very real consequences. But journal articles, a primary way science is communicated in academia, are a different format to newspaper articles or blogs and require a level of skill and undoubtedly a greater amount of patience. Here Jennifer Raff has prepared a helpful guide for non-scientists on how to read a scientific paper. These steps and tips will be useful to anyone interested in the presentation of scientific findings and raise important points for scientists to consider with their own writing practice.
My post, The truth about vaccinations: Your physician knows more than the University of Google sparked a very lively discussion, with comments from several people trying to persuade me (and the other readers) that their paper disproved everything that I’d been saying. While I encourage you to go read the comments and contribute your own, here I want to focus on the much larger issue that this debate raised: what constitutes scientific authority?
It’s not just a fun academic problem. Getting the science wrong has very real consequences. For example, when a community doesn’t vaccinate children because they’re afraid of “toxins” and think that prayer (or diet, exercise, and “clean living”) is enough to prevent infection, outbreaks happen.
“Be skeptical. But when you get proof, accept proof.” –Michael Specter
What constitutes enough proof? Obviously everyone has a different answer to that question. But to form a truly educated opinion on a scientific subject, you need to become familiar with current research in that field. And to do that, you have to read the “primary research literature” (often just called “the literature”). You might have tried to read scientific papers before and been frustrated by the dense, stilted writing and the unfamiliar jargon. I remember feeling this way! Reading and understanding research papers is a skill which every single doctor and scientist has had to learn during graduate school. You can learn it too, but like any skill it takes patience and practice.
I want to help people become more scientifically literate, so I wrote this guide for how a layperson can approach reading and understanding a scientific research paper. It’s appropriate for someone who has no background whatsoever in science or medicine, and based on the assumption that he or she is doing this for the purpose of getting a basic understanding of a paper and deciding whether or not it’s a reputable study.
The type of scientific paper I’m discussing here is referred to as a primary research article . It’s a peer-reviewed report of new research on a specific question (or questions). Another useful type of publication is a review article . Review articles are also peer-reviewed, and don’t present new information, but summarize multiple primary research articles, to give a sense of the consensus, debates, and unanswered questions within a field. (I’m not going to say much more about them here, but be cautious about which review articles you read. Remember that they are only a snapshot of the research at the time they are published. A review article on, say, genome-wide association studies from 2001 is not going to be very informative in 2013. So much research has been done in the intervening years that the field has changed considerably).
Before you begin: some general advice
Reading a scientific paper is a completely different process than reading an article about science in a blog or newspaper. Not only do you read the sections in a different order than they’re presented, but you also have to take notes, read it multiple times, and probably go look up other papers for some of the details. Reading a single paper may take you a very long time at first. Be patient with yourself. The process will go much faster as you gain experience.
Most primary research papers will be divided into the following sections: Abstract, Introduction, Methods, Results, and Conclusions/Interpretations/Discussion. The order will depend on which journal it’s published in. Some journals have additional files (called Supplementary Online Information) which contain important details of the research, but are published online instead of in the article itself (make sure you don’t skip these files).
Before you begin reading, take note of the authors and their institutional affiliations. Some institutions (e.g. University of Texas) are well-respected; others (e.g. the Discovery Institute ) may appear to be legitimate research institutions but are actually agenda-driven. Tip: g oogle “Discovery Institute” to see why you don’t want to use it as a scientific authority on evolutionary theory.
Also take note of the journal in which it’s published. Reputable (biomedical) journals will be indexed by Pubmed . [EDIT: Several people have reminded me that non-biomedical journals won’t be on Pubmed, and they’re absolutely correct! (thanks for catching that, I apologize for being sloppy here). Check out Web of Science for a more complete index of science journals. And please feel free to share other resources in the comments!] Beware of questionable journals .
As you read, write down every single word that you don’t understand. You’re going to have to look them all up (yes, every one. I know it’s a total pain. But you won’t understand the paper if you don’t understand the vocabulary. Scientific words have extremely precise meanings).
Step-by-step instructions for reading a primary research article
1. Begin by reading the introduction, not the abstract.
The abstract is that dense first paragraph at the very beginning of a paper. In fact, that’s often the only part of a paper that many non-scientists read when they’re trying to build a scientific argument. (This is a terrible practice—don’t do it.). When I’m choosing papers to read, I decide what’s relevant to my interests based on a combination of the title and abstract. But when I’ve got a collection of papers assembled for deep reading, I always read the abstract last. I do this because abstracts contain a succinct summary of the entire paper, and I’m concerned about inadvertently becoming biased by the authors’ interpretation of the results.
2. Identify the BIG QUESTION.
Not “What is this paper about”, but “What problem is this entire field trying to solve?”
This helps you focus on why this research is being done. Look closely for evidence of agenda-motivated research.
3. Summarize the background in five sentences or less.
Here are some questions to guide you:
What work has been done before in this field to answer the BIG QUESTION? What are the limitations of that work? What, according to the authors, needs to be done next?
The five sentences part is a little arbitrary, but it forces you to be concise and really think about the context of this research. You need to be able to explain why this research has been done in order to understand it.
4. Identify the SPECIFIC QUESTION(S)
What exactly are the authors trying to answer with their research? There may be multiple questions, or just one. Write them down. If it’s the kind of research that tests one or more null hypotheses, identify it/them.
Not sure what a null hypothesis is? Go read this one and try to identify the null hypotheses in it. Keep in mind that not every paper will test a null hypothesis.
5. Identify the approach
What are the authors going to do to answer the SPECIFIC QUESTION(S)?
6. Now read the methods section. Draw a diagram for each experiment, showing exactly what the authors did.
I mean literally draw it. Include as much detail as you need to fully understand the work. As an example, here is what I drew to sort out the methods for a paper I read today ( Battaglia et al. 2013: “The first peopling of South America: New evidence from Y-chromosome haplogroup Q” ). This is much less detail than you’d probably need, because it’s a paper in my specialty and I use these methods all the time. But if you were reading this, and didn’t happen to know what “process data with reduced-median method using Network” means, you’d need to look that up.
Image credit: author
You don’t need to understand the methods in enough detail to replicate the experiment—that’s something reviewers have to do—but you’re not ready to move on to the results until you can explain the basics of the methods to someone else.
7. Read the results section. Write one or more paragraphs to summarize the results for each experiment, each figure, and each table. Don’t yet try to decide what the results mean , just write down what they are.
You’ll find that, particularly in good papers, the majority of the results are summarized in the figures and tables. Pay careful attention to them! You may also need to go to the Supplementary Online Information file to find some of the results.
It is at this point where difficulties can arise if statistical tests are employed in the paper and you don’t have enough of a background to understand them. I can’t teach you stats in this post, but here , and here are some basic resources to help you. I STRONGLY advise you to become familiar with them.
Things to pay attention to in the results section:
- Any time the words “significant” or “non-significant” are used. These have precise statistical meanings. Read more about this here .
- If there are graphs, do they have error bars on them? For certain types of studies, a lack of confidence intervals is a major red flag.
- The sample size. Has the study been conducted on 10, or 10,000 people? (For some research purposes, a sample size of 10 is sufficient, but for most studies larger is better).
8. Do the results answer the SPECIFIC QUESTION(S)? What do you think they mean?
Don’t move on until you have thought about this. It’s okay to change your mind in light of the authors’ interpretation—in fact you probably will if you’re still a beginner at this kind of analysis—but it’s a really good habit to start forming your own interpretations before you read those of others.
9. Read the conclusion/discussion/Interpretation section.
What do the authors think the results mean? Do you agree with them? Can you come up with any alternative way of interpreting them? Do the authors identify any weaknesses in their own study? Do you see any that the authors missed? (Don’t assume they’re infallible!) What do they propose to do as a next step? Do you agree with that?
10. Now, go back to the beginning and read the abstract.
Does it match what the authors said in the paper? Does it fit with your interpretation of the paper?
11. FINAL STEP: (Don’t neglect doing this) What do other researchers say about this paper?
Who are the (acknowledged or self-proclaimed) experts in this particular field? Do they have criticisms of the study that you haven’t thought of, or do they generally support it?
Here’s a place where I do recommend you use google! But do it last, so you are better prepared to think critically about what other people say.
(12. This step may be optional for you, depending on why you’re reading a particular paper. But for me, it’s critical! I go through the “Literature cited” section to see what other papers the authors cited. This allows me to better identify the important papers in a particular field, see if the authors cited my own papers (KIDDING!….mostly), and find sources of useful ideas or techniques.)
UPDATE: If you would like to see an example of how to read a science paper using this framework, you can find one here .
I gratefully acknowledge Professors José Bonner and Bill Saxton for teaching me how to critically read and analyze scientific papers using this method. I’m honored to have the chance to pass along what they taught me.
I’ve written a shorter version of this guide for teachers to hand out to their classes. If you’d like a PDF, shoot me an email: jenniferraff (at) utexas (dot) edu. For further comments and additional questions on this guide, please see the Comments Section on the original post .
This piece originally appeared on the author’s personal blog and is reposted with permission.
Featured image credit: Scientists in a laboratory of the University of La Rioja by Urcomunicacion (Wikimedia CC BY3.0)
Note: This article gives the views of the authors, and not the position of the LSE Impact blog, nor of the London School of Economics. Please review our Comments Policy if you have any concerns on posting a comment below.
Jennifer Raff (Indiana University—dual Ph.D. in genetics and bioanthropology) is an assistant professor in the Department of Anthropology, University of Kansas, director and Principal Investigator of the KU Laboratory of Human Population Genomics, and assistant director of KU’s Laboratory of Biological Anthropology. She is also a research affiliate with the University of Texas anthropological genetics laboratory. She is keenly interested in public outreach and scientific literacy, writing about topics in science and pseudoscience for her blog ( violentmetaphors.com ), the Huffington Post, and for the Social Evolution Forum .
- Learning Consultations
- Peer Tutoring
- Getting Started
- Peer Education Courses
- Become a Peer Educator
- ADHD/LD Support
- Workshops & Outreach
- Learning Strategies
- Manage Time
- All Resources
- For Faculty & Staff
Reference management. Clean and simple.
How to read a scientific paper: a step-by-step guide

Research Process
- Brainstorming
- Explore Google This link opens in a new window
- Explore Web Resources
- Explore Background Information
- Explore Books
- Explore Scholarly Articles
- Narrowing a Topic
- Primary and Secondary Resources
- Academic, Popular & Trade Publications
- Scholarly and Peer-Reviewed Journals
- Grey Literature
- Clinical Trials
- Evidence Based Treatment
- Scholarly Research
- Database Research Log
- Search Limits
- Keyword Searching
- Boolean Operators
- Phrase Searching
- Truncation & Wildcard Symbols
- Proximity Searching
- Field Codes
- Subject Terms and Database Thesauri
Reading a Scientific Article
- Website Evaluation
- Article Keywords and Subject Terms
- Cited References
- Citing Articles
- Related Results
- Search Within Publication
- Database Alerts & RSS Feeds
- Personal Database Accounts
- Persistent URLs
- Literature Gap and Future Research
- Web of Knowledge
- Annual Reviews
- Systematic Reviews & Meta-Analyses
- Finding Seminal Works
- Exhausting the Literature
- Finding Dissertations
- Researching Theoretical Frameworks
- Research Methodology & Design
- Tests and Measurements
- Organizing Research & Citations This link opens in a new window
- Scholarly Publication
- Learn the Library This link opens in a new window
Library Tutorial
- Reading a Scholarly Article Tutorial This interactive tutorial provides practice reading a scholarly or scientific article.
Additional Resources
- Anatomy of a Scholarly Article
- How to Read (and Understand) a Social Science Journal Article
- How to Read a Scientific Paper
- How to Read a Scientific Paper Interactive Tutorial
- How to Read Scientific Literature (YouTube Video)
General Dictionaries
- The American Heritage Dictionary of the English Language
- The American Heritage Student Science Dictionary
- The Chambers Dictionary
- Dictionary.com
- The Free Dictionary
- Merriam-Webster's Collegiate Dictionary
- Merriam-Webster Online
- The Penguin English Dictionary
- The Science Dictionary
Attempting to read a scientific or scholarly research article for the first time may seem overwhelming and confusing. This guide details how to read a scientific article step-by-step. First, you should not approach a scientific article like a textbook— reading from beginning to end of the chapter or book without pause for reflection or criticism. Additionally, it is highly recommended that you highlight and take notes as you move through the article. Taking notes will keep you focused on the task at hand and help you work towards comprehension of the entire article.
- Skim the article. This should only take you a few minutes. You are not trying to comprehend the entire article at this point, but just get a basic overview. You don’t have to read in order; the discussion/conclusions will help you to determine if the article is relevant to your research. You might then continue on to the Introduction. Pay attention to the structure of the article, headings, and figures.
- Grasp the vocabulary. Begin to go through the article and highlight words and phrases you do not understand. Some words or phrases you may be able to get an understanding from the context in which it is used, but for others you may need the assistance of a medical or scientific dictionary. Subject-specific dictionaries available through our Library databases and online are listed below.
- The abstract gives a quick overview of the article. It will usually contain four pieces of information: purpose or rationale of study (why they did it); methodology (how they did it); results (what they found); conclusion (what it means). Begin by reading the abstract to make sure this is what you are looking for and that it will be worth your time and effort.
- The introduction gives background information about the topic and sets out specific questions to be addressed by the authors. You can skim through the introduction if you are already familiar with the paper’s topic.
- The methods section gives technical details of how the experiments were carried out and serves as a “how-to” manual if you wanted to replicate the same experiments as the authors. This is another section you may want to only skim unless you wish to identify the methods used by the researchers or if you intend to replicate the research yourself.
- The results are the meat of the scientific article and contain all of the data from the experiments. You should spend time looking at all the graphs, pictures, and tables as these figures will contain most of the data.
- Lastly, the discussion is the authors’ opportunity to give their opinions. Keep in mind that the discussions are the authors’ interpretations and not necessarily facts. It is still a good place for you to get ideas about what kind of research questions are still unanswered in the field and what types of questions you might want your own research project to tackle. (See the Future Research Section of the Research Process for more information).
- Read the bibliography/references section. Reading the references or works cited may lead you to other useful resources. You might also get a better understanding of the basic terminology, main concepts, major researchers, and basic terminology in the area you are researching.
- Have I taken time to understand all the terminology?
- Am I spending too much time on the less important parts of this article?
- Do I have any reason to question the credibility of this research?
- What specific problem does the research address and why is it important?
- How do these results relate to my research interests or to other works which I have read?
- Read the article a second time in chronological order. Reading the article a second time will reinforce your overall understanding. You may even start to make connections to other articles that you have read on this topic.
Reading a Scholarly Article Workshop
This workshop presents effective techniques for reading and understanding a scholarly article, as well as locating definitions related to your research topic.
Subject-Specific Dictionaries
- Health Sciences
- Marriage & Family Science
- Research Methods
- Social Work
Was this resource helpful?
- << Previous: Subject Terms and Database Thesauri
- Next: Evaluating Information >>
- Last Updated: Mar 31, 2024 4:57 PM
- URL: https://resources.nu.edu/researchprocess

© Copyright 2024 National University. All Rights Reserved.
Privacy Policy | Consumer Information
- How To Find Articles with Databases
- How To Evaluate Articles
- How To Read A Scientific Paper
- How To Interpret Data
- How To Write A Lab Report
- How To Write A Scientific Paper
- Get More Help
- Reference: Encyclopedia, Handbooks & Dictionaries
- Research Tools: Databases, Protocols & Citation Locators
- E-Journal Lists by Subject
- Scholarly vs Popular
- Search Tips
- Open Resources
- E-Journal lists by subject
- Develop a Research Question
Useful Sources
- How to (Seriously) Read a Scientific Paper
- How to Read a Scientific Article
- Infographic: How to Read a Scientific Paper
Reading a Scientific Paper
Reading a scientific paper can seem like a daunting task. However, learning how to properly read a scholarly article can make the process much easier! Understanding the different parts of a scientific article can help the reader to understand the material.
- The title of the article can give the reader a lot of information about its contents, such as the topic, major ideas, and participants.
- Abstracts help to summarize the article and give the reader a preview of the material they are about to read. The abstract is very important and should be read with care.
Introduction
- What is the article's purpose being stated in the introduction?
- Why would this article be of interest to experts in the field?
- What is already known, or not known, about this topic?
- What specifically is the hypothesis? If one is not given, what are the expectations of the author?
- Having these questions in mind when reading the introduction can help the reader gain an understanding of the article as a whole. A good research article will answer these questions in the introduction and be consistent with their explanation throughout the rest of the article.
- What are the specific methods used by the researcher?
- Does the researcher provide a coherent and viable plan for their experiment?
- Has the author missed any variables that could effect the results of their findings?
- How do the methods in this article compare with similar articles?
- Ex: they are correlated and support the hypothesis, they contradict they hypothesis, ect.
- If there are differences from the hypothesis, what differences did the researcher find?
- Are the findings described in an unbiased way?
- Is there new information presented that wasn't known before?
- Is the researcher unbiased in their presentation?
- Ex: More research needs to be done, the findings show a solution to a known problem, etc.
- What suggestions are made about future research? If no suggestions are made, should there be?
- The conclusion points out the important findings from the experiment or research. Occasionally, it will incorporated into the discussion section of the paper.
General Tips
- Fully comprehending a scientific article will most likely take more than one read. Don't be discouraged if you don't understand everything the first time, reading scientific papers is a skill that is developed with practice.
- Start with the broad and then to the specific. Begin by understanding the topic of the article before trying to dig through all the fine points the author is making.
- Always read the tables, charts, and figures. These will give a visual clue to the methods and results sections of the paper and help you to understand the data. The author put these in the paper for a reason, don't dismiss their importance.
- Don't be afraid to ask questions or look up definitions. If you do not understand a term or concept, do not be afraid to ask for help or look up an explanation.
- << Previous: How To Evaluate Articles
- Next: How To Interpret Data >>
- Last Updated: Mar 8, 2024 2:26 PM
- URL: https://guides.libraries.indiana.edu/STEM
Social media
- Instagram for Herman B Wells Library
- Facebook for IU Libraries
Additional resources
Featured databases.
- Resource available to authorized IU Bloomington users (on or off campus) OneSearch@IU
- Resource available to authorized IU Bloomington users (on or off campus) Academic Search (EBSCO)
- Resource available to authorized IU Bloomington users (on or off campus) ERIC (EBSCO)
- Resource available to authorized IU Bloomington users (on or off campus) Nexis Uni
- Resource available without restriction HathiTrust Digital Library
- Databases A-Z
- Resource available to authorized IU Bloomington users (on or off campus) Google Scholar
- Resource available to authorized IU Bloomington users (on or off campus) JSTOR
- Resource available to authorized IU Bloomington users (on or off campus) Web of Science
- Resource available to authorized IU Bloomington users (on or off campus) Scopus
- Resource available to authorized IU Bloomington users (on or off campus) WorldCat
IU Libraries
- Diversity Resources
- About IU Libraries
- Alumni & Friends
- Departments & Staff
- Jobs & Libraries HR
- Intranet (Staff)
- IUL site admin

Research Techniques for Undergraduate Research
- Library Research
- Citation Tracing/Tracking in Google Scholar
- Strategies for Research
- Chicago Manual of Style
- Writing Research Papers
- How to Read a Citation
- How to read and understand a scientific paper
- Skimming an article
- Workshop presentation powerpoint
- Post-Workshop Quiz
- Post-Workshop Reflection
How to Read a Scientific Paper overview
Below, you'll find two different articles about how to read a scientific paper. The second one is written by a science journalist and was added to this guide in 2024. We hope you find both articles useful. They overlap and bring useful techniques to light.
How to read and understand a scientific paper: a guide for non-scientists
- Handout for How to Read and Understand a Scientific Paper: A Guide for Non-Scientists
Reprinted by permission of the author, Jennifer Raff, Assistant Professor, Department of Anthropology, University of Kansas, https://about.me/jenniferraff :: original URL: https://violentmetaphors.com/2013/08/25/how-to-read-and-understand-a-scientific-paper-2/ Last week’s post ( The truth about vaccinations: Your physician knows more than the University of Google ) sparked a very lively discussion, with comments from several people trying to persuade me (and the other readers) that their paper disproved everything that I’d been saying. While I encourage you to go read the comments and contribute your own, here I want to focus on the much larger issue that this debate raised: what constitutes scientific authority?
It’s not just a fun academic problem. Getting the science wrong has very real consequences. For example, when a community doesn’t vaccinate children because they’re afraid of “toxins” and think that prayer (or diet, exercise, and “clean living”) is enough to prevent infection, outbreaks happen .
“Be skeptical. But when you get proof, accept proof.” –Michael Specter
What constitutes enough proof? Obviously everyone has a different answer to that question. But to form a truly educated opinion on a scientific subject, you need to become familiar with current research in that field. And to do that, you have to read the “primary research literature” (often just called “the literature”). You might have tried to read scientific papers before and been frustrated by the dense, stilted writing and the unfamiliar jargon. I remember feeling this way! Reading and understanding research papers is a skill which every single doctor and scientist has had to learn during graduate school. You can learn it too, but like any skill it takes patience and practice.
I want to help people become more scientifically literate, so I wrote this guide for how a layperson can approach reading and understanding a scientific research paper. It’s appropriate for someone who has no background whatsoever in science or medicine, and based on the assumption that he or she is doing this for the purpose of getting a basic understanding of a paper and deciding whether or not it’s a reputable study.
The type of scientific paper I’m discussing here is referred to as a primary research article . It’s a peer-reviewed report of new research on a specific question (or questions). Another useful type of publication is a review article . Review articles are also peer-reviewed, and don’t present new information, but summarize multiple primary research articles, to give a sense of the consensus, debates, and unanswered questions within a field. (I’m not going to say much more about them here, but be cautious about which review articles you read. Remember that they are only a snapshot of the research at the time they are published. A review article on, say, genome-wide association studies from 2001 is not going to be very informative in 2013. So much research has been done in the intervening years that the field has changed considerably).
Before you begin: some general advice Reading a scientific paper is a completely different process than reading an article about science in a blog or newspaper. Not only do you read the sections in a different order than they’re presented, but you also have to take notes, read it multiple times, and probably go look up other papers for some of the details. Reading a single paper may take you a very long time at first. Be patient with yourself. The process will go much faster as you gain experience.
Most primary research papers will be divided into the following sections: Abstract, Introduction, Methods, Results, and Conclusions/Interpretations/Discussion. The order will depend on which journal it’s published in. Some journals have additional files (called Supplementary Online Information) which contain important details of the research, but are published online instead of in the article itself (make sure you don’t skip these files).
Before you begin reading, take note of the authors and their institutional affiliations. Some institutions (e.g. University of Texas) are well-respected; others (e.g. the Discovery Institute ) may appear to be legitimate research institutions but are actually agenda-driven. Tip: g oogle “Discovery Institute” to see why you don’t want to use it as a scientific authority on evolutionary theory.
Also take note of the journal in which it’s published. Reputable (biomedical) journals will be indexed by Pubmed . [ EDIT: Several people have reminded me that non-biomedical journals won’t be on Pubmed, and they’re absolutely correct! (thanks for catching that, I apologize for being sloppy here). Check out Web of Science for a more complete index of science journals. And please feel free to share other resources in the comments!] Beware of questionable journals .
As you read, write down every single word that you don’t understand. You’re going to have to look them all up (yes, every one. I know it’s a total pain. But you won’t understand the paper if you don’t understand the vocabulary. Scientific words have extremely precise meanings).
Step-by-step instructions for reading a primary research article
1. Begin by reading the introduction, not the abstract.
The abstract is that dense first paragraph at the very beginning of a paper. In fact, that’s often the only part of a paper that many non-scientists read when they’re trying to build a scientific argument. (This is a terrible practice—don’t do it.). When I’m choosing papers to read, I decide what’s relevant to my interests based on a combination of the title and abstract. But when I’ve got a collection of papers assembled for deep reading, I always read the abstract last . I do this because abstracts contain a succinct summary of the entire paper, and I’m concerned about inadvertently becoming biased by the authors’ interpretation of the results.
2. Identify the BIG QUESTION.
Not “What is this paper about”, but “What problem is this entire field trying to solve?”
This helps you focus on why this research is being done. Look closely for evidence of agenda-motivated research.
3. Summarize the background in five sentences or less.
Here are some questions to guide you:
What work has been done before in this field to answer the BIG QUESTION? What are the limitations of that work? What, according to the authors, needs to be done next?
The five sentences part is a little arbitrary, but it forces you to be concise and really think about the context of this research. You need to be able to explain why this research has been done in order to understand it.
4. Identify the SPECIFIC QUESTION(S)
What exactly are the authors trying to answer with their research? There may be multiple questions, or just one. Write them down. If it’s the kind of research that tests one or more null hypotheses, identify it/them.
Not sure what a null hypothesis is? Go read this , then go back to my last post and read one of the papers that I linked to (like this one ) and try to identify the null hypotheses in it. Keep in mind that not every paper will test a null hypothesis.
5. Identify the approach
What are the authors going to do to answer the SPECIFIC QUESTION(S)?
6. Now read the methods section. Draw a diagram for each experiment, showing exactly what the authors did.
I mean literally draw it. Include as much detail as you need to fully understand the work. As an example, here is what I drew to sort out the methods for a paper I read today ( Battaglia et al. 2013: “The first peopling of South America: New evidence from Y-chromosome haplogroup Q” ). This is much less detail than you’d probably need, because it’s a paper in my specialty and I use these methods all the time. But if you were reading this, and didn’t happen to know what “process data with reduced-median method using Network” means, you’d need to look that up.

You don’t need to understand the methods in enough detail to replicate the experiment—that’s something reviewers have to do—but you’re not ready to move on to the results until you can explain the basics of the methods to someone else.
7. Read the results section. Write one or more paragraphs to summarize the results for each experiment, each figure, and each table. Don’t yet try to decide what the results mean , just write down what they are.
You’ll find that, particularly in good papers, the majority of the results are summarized in the figures and tables. Pay careful attention to them! You may also need to go to the Supplementary Online Information file to find some of the results.
It is at this point where difficulties can arise if statistical tests are employed in the paper and you don’t have enough of a background to understand them. I can’t teach you stats in this post, but here , here , and here are some basic resources to help you. I STRONGLY advise you to become familiar with them.
THINGS TO PAY ATTENTION TO IN THE RESULTS SECTION:
-Any time the words “ significant ” or “ non-significant ” are used. These have precise statistical meanings. Read more about this here .
-If there are graphs, do they have error bars on them? For certain types of studies, a lack of confidence intervals is a major red flag.
-The sample size. Has the study been conducted on 10, or 10,000 people? (For some research purposes, a sample size of 10 is sufficient, but for most studies larger is better).
8. Do the results answer the SPECIFIC QUESTION(S)? What do you think they mean?
Don’t move on until you have thought about this. It’s okay to change your mind in light of the authors’ interpretation—in fact you probably will if you’re still a beginner at this kind of analysis—but it’s a really good habit to start forming your own interpretations before you read those of others.
9. Read the conclusion/discussion/Interpretation section.
What do the authors think the results mean? Do you agree with them? Can you come up with any alternative way of interpreting them? Do the authors identify any weaknesses in their own study? Do you see any that the authors missed? (Don’t assume they’re infallible!) What do they propose to do as a next step? Do you agree with that?
10. Now, go back to the beginning and read the abstract.
Does it match what the authors said in the paper? Does it fit with your interpretation of the paper?
11. FINAL STEP: (Don’t neglect doing this) What do other researchers say about this paper?
Who are the (acknowledged or self-proclaimed) experts in this particular field? Do they have criticisms of the study that you haven’t thought of, or do they generally support it?
Here’s a place where I do recommend you use google! But do it last, so you are better prepared to think critically about what other people say.
(12. This step may be optional for you, depending on why you’re reading a particular paper. But for me, it’s critical! I go through the “Literature cited” section to see what other papers the authors cited. This allows me to better identify the important papers in a particular field, see if the authors cited my own papers (KIDDING!….mostly), and find sources of useful ideas or techniques.)
Now brace for more conflict– next week we’re going to use this method to go through a paper on a controversial subject! Which one would you like to do? Shall we critique one of the papers I posted last week?
UPDATE: If you would like to see an example, you can find one here ———————————————————————————————————
I gratefully acknowledge Professors José Bonner and Bill Saxton for teaching me how to critically read and analyze scientific papers using this method. I’m honored to have the chance to pass along what they taught me.
How to Read a Scientific Paper by a science journalist
How to read a scientific paper.
- Alexandra Witze
- November 6, 2018
Léelo en español

It’s one of the first, and likely most intimidating, assignments for a fledgling science reporter. “Here,” your editor says. “Write up this paper that’s coming out in Science this week.” And suddenly you’re staring at an impenetrable PDF—pages of scientific jargon that you’re supposed to understand, interview the author and outside commenters about, and describe in ordinary English to ordinary readers.
Fear not! The Open Notebook is here with a primer on how to read a scientific paper. These tips and tricks will work whether you’re covering developmental biology or deep-space exploration. The key is to familiarize yourself with the framework in which scientists describe their discoveries, and to not let yourself get bogged down in detail as you’re trying to understand the overarching point of it all. As a specific example, we’ve marked up a Science paper in the accompanying image.
But first, let’s break down what a typical scientific paper contains. Most include these basic sections, usually in this order:
The author list is as it sounds, a roster of the scientists involved in the discovery. But hidden within the names are clues that will help you navigate the politics of reporting the story. The first name in the list is often (but not always) the person who did the most work, perhaps the graduate student or postdoc who is the lead on the project. This person is usually (but not always) designated as the “corresponding author” by an asterisk by their name, or by their email address being given on the first or last page of the paper. If the corresponding author is not the first name in the author list, then take extra care to Google the various authors and figure out how they relate to one another. (In many fields, such as biology and psychology, the last author in the list is typically the senior author or lab head. In others, such as experimental physics where the author list can number in the dozens or hundreds, authors are usually listed alphabetically.) The senior author might be able to provide some broad perspective as to why and how the study was undertaken. But the first or corresponding author is much more likely to be the person who actually did the work, and therefore your better request for an interview.
The abstract is a summary of the paper’s conclusions. Always read this first, several times over. Usually the significance of the paper will be laid out here, albeit in technical terms. A good abstract will summarize what research was undertaken, what the scientists found, and why it’s important. (Compare the abstract of this recent Nature paper , on the discovery of a prehistoric human hybrid, to the first three paragraphs of Sarah Kaplan’s Washington Post story reporting the discovery . Kaplan clearly captures the essence of the new findings as described in the abstract.) Relevant numbers such as the statistical significance of the finding are often highlighted here as well. Abstracts are prone to typographical errors, so be sure to double-check numbers against the body of the paper as well as your interview with the author.
The body of the paper lays out the bulk of the scientific findings. Pay special attention to the first couple of paragraphs, which often serve as an introduction, describing previous research in the field and why the new work is important. This is an excellent place to hunt for references to other papers that can serve as your guidepost for outside commenters (more on that later). Next will come the details of how the research was done; sometimes much of this is broken out into a later methods section (see below). Then come the results , which may be lengthy. Look for phrases such as “we concluded” to clue you in to their most important points. If statistics are involved, see Rachel Zamzow’s primer on how to spot shady statistics.
The final section (sometimes labeled as discussion ) often summarizes the new findings, puts them in context, and describes the likely next steps to be taken. If your reading has been dragging through the results section, now is the time to refocus. “That sort of information will help a writer answer the nearly inevitable “so what?” question for their readers as well as their editors,” says Sid Perkins, a freelance science writer in Crossville, Tennessee, who writes for outlets including Science and Science News for Students .
The figures are the data, graphics, or other visual representations of the discovery. Read these and their captions carefully, as they often contain the bulk of the new findings. If you don’t understand the figures, ask the scientist to walk you through them during your interview. Don’t be afraid to say things like, “I don’t understand what the x-axis means.”
The references are your portal into a world of additional inscrutable PDFs. You need to plow through at least a couple of the citations, because they are your initial guide in figuring out who you need to call for outside comment. The references are referenced (usually by number) within the body of the text, so you can pinpoint the ones that will be most helpful. For instance, if the text talks about how previous studies have found the opposite of this new one, go look up the cited references, because those authors would be excellent outside commenters. If you do not have access to the journals described in the references, you can at least look at the paper abstract, which is always outside the paywall , to get a sense of what those earlier studies concluded. (For further caveats on references, see below.)
The acknowledgments are meant for transparency, to show the contributions of the various authors and where they got their funding from. Things to look for in here are whether they thank other scientists for “discussions” or “review” of the work; sometimes peer reviewers are explicitly acknowledged as such, in which case you can call those people right away for outside comment. Occasionally there are humorous tidbits that you can pick up on for a story , such as when authors thank the field-camp guards who kept them safe from predatory polar bears . The funding section is usually pro forma, but it is worth scanning for mention of unusual sources of income, such as from a science-loving philanthropist. If the authors declare competing financial interests (such as a patent filing) you will need to report those out and make sure you understand what financial conflicts of interest may be clouding their objectivity.
The methods often appear in a ridiculously small typeface after the body of the paper. These lay out how the actual experiments were done. Scour these for any details that will bring your story to life. For instance, they might describe how the climate models were so complicated that they took more than a year to run on one of the world’s most powerful supercomputers.
Supplementary information comes with some but not all papers. In most cases it is extra material that the journal did not want to devote space to describing in the paper itself. Always check it out, because there may be hidden gems. In a 2015 study of global lake warming , the only way to find out which specific lakes were warming—and talk about the nearest ones for readers —was to wade through the supplementary information. In another recent example, Harvard researchers left it to the supplementary information to explain that they cranked up a leaf-blower to see how lizards fared during hurricanes, a fact that the Associated Press’s Seth Borenstein turned into his lede .
So now you’re armed with the basics of what makes up a science paper. How should you tackle reading for your next assignment? The task will be more manageable if you break it into a series of jobs.
Strategize During the First Pass
Your first dive into a paper should be aimed at gathering the most important information for your story—that is, what the research found and why anyone should care. For that, consider following the approach of Mark Peplow, a freelance science journalist in Cambridge, England, who writes for publications including Nature and Chemical & Engineering News .
If it’s a field he’s relatively familiar with, such as chemistry or materials science, Peplow takes a first pass through the paper, underlining with a red pen all the facts that are likely to make it into his initial draft. “That means I can produce a skeleton first draft of the story by simply writing a series of sentences containing what I’ve underlined, and then go into editing mode to jigsaw them into the right order,” he says. (In my annotated example, I’ve done this for the abstract using a purple pen.)

As Peplow reads, he looks for numbers to help make the story sing (“… so porous that a chunk of material the size of a sugar cube contains the surface area of 17 tennis courts”—see orange highlighter in the annotated paper) and methodological details that might prompt a fun interview question (“How scary was it to be pouring that very hazardous liquid into another one?”). He also keeps an eye out for anything indicating an emerging trend or other examples of the same phenomenon, which can be useful for context within the story or as a forward-looking kicker (see how he pulls this off in this Chemical & Engineering News story) .
But what if the paper is in a field you’re not experienced with, and you don’t understand the terminology? Peplow has a plan for that too. “I read the abstract, bathe in my lack of understanding, and mentally throw the abstract away,” he says.
Then he goes through the paper, underlining fragments he understands and putting wiggly lines next to paragraphs that he thinks sound important, but doesn’t actually know what they mean. Jargon words get circled, and equations ignored. He forges onward, paying attention to phrases such as “our findings,” “revealed,” “established,” or “our measurements show”—signs that these are the new and important bits. “Once I’ve reached the end of the paper, and I’m sure I don’t understand it, I remind myself it’s not my fault,” Peplow says.
At that point, Peplow starts looking up definitions for the jargon words, either with Google or Wikipedia or in a stack of science reference books he picked up for free when a local library closed. He jots definitions of the words on the paper. To understand concepts, he sometimes searches EurekAlert! for past press releases that explain core concepts, or Googles a string of keywords and adds “review” to hunt for a more comprehensible description.
By this point, Peplow can circle back to the paragraphs marked with wiggly lines and start to understand them better. What he doesn’t yet comprehend, he marks down as an interview question for the researcher.
Circle Back for What You May Have Missed
Before picking up the phone for that interview, it’s worth making a second pass through the paper to see what else you need to help you in your reporting. Check, usually near the end of the paper, to see whether the scientists discuss what the next steps should be—either for their own team or for other groups following up to confirm or expand on the new results, says Perkins. That can provide a ready-made kicker for your story.
Susan Milius, a reporter who covers the life sciences for Science News , often makes a beeline straight for the references to try to start identifying outside commenters for a piece. She will find those PDFs and then look within the references’ references to build a broad understanding of the field. One caveat, though: Be sure to research how these possible commenters are connected to the author of the current study. Once, Milius phoned an outside commenter who had published on the topic in question some years earlier—but that scientist turned out to be the spouse of the new paper’s author. She had a different last name than her husband.
It’s also worth remembering that the authors may well be biased in which references they include in the paper. Self-citations, in which authors try to boost their citation count by adding their previous publications to the reference list, are common. And sometimes authors deliberately omit papers by competing groups, a fact that is not always caught during the peer-review process. So don’t rely on the references within the PDF to be comprehensive; try a Google Scholar search using keywords from the paper to unearth whether there are competing groups out there.
Other clues may lie in how long the manuscript took to make it through the peer-review process. For many journals these dates come at the very end of the paper, marked something like “submitted” and “accepted.” Different journals have different timescales for publishing, but it is always worth looking to see whether the manuscript languished an extraordinary amount of time (like many months) in the review process. If so, ask the author why things took so long. (A fairly innocuous way to do this is to say something like, “I noticed it took a while for this paper to be accepted. Can you tell me how that process went?” Then be prepared for the authors to go on a rant about peer review.)
Hunt for Extra Details
Finally, see if there are additional sources of information you can sweep into your reporting. Check to see if the author’s institution is issuing a press release about the work; if this isn’t already posted on EurekAlert!, ask the author during the interview if they are preparing additional press materials and, if so, how you can get hold of those. This is also a good time to ask for any art, such as photos or videos to illustrate your story. You will of course have already looked at all their figures in detail, so you’ll be well placed to request the art that is most relevant to what you and your editor are looking for.
With these tools at your side, you should be well suited to tackle your next scientific paper.

Alexandra Witze is a science journalist in Boulder, Colorado, and a member of The Open Notebook ’s board of directors. Her news story on the Martian subglacial lake (marked up above) appeared in Nature . Follow her on
- << Previous: How to Read a Citation
- Next: Skimming an article >>
- Last Updated: Mar 27, 2024 12:30 PM
- URL: https://libguides.citytech.cuny.edu/advancedResearch
How to Read a Scientific Paper

To read a scientific paper effectively, you should focus on the results and ensure that you draw your own conclusions from the data and assess whether this agrees with the authors’ conclusions. You should also check that the methods are appropriate and make sense. Spend time attending journal clubs and reading online peer reviews of articles to help hone your critical analysis skills and make reading papers easier and quicker.
Keeping up with the scientific literature in your field of interest is incredibly important. It keeps you informed about what is happening in your field and helps shape and guide your experimental plans. But do you really know how to read a scientific paper, and can you do it effectively and efficiently?
Let’s face it, in our results-driven world, reading new scientific papers often falls by the wayside because we just don’t have the time! And when you do find some reading time, it’s tempting not to read the entire article and just focus on the abstract and conclusions sections.
But reading a scientific paper properly doesn’t need to take hours of your time. We’ll show you how to read a scientific paper effectively, what you can and can’t skim, and give you a checklist of key points to look for when reading a paper to make sure you get the most out of your time.
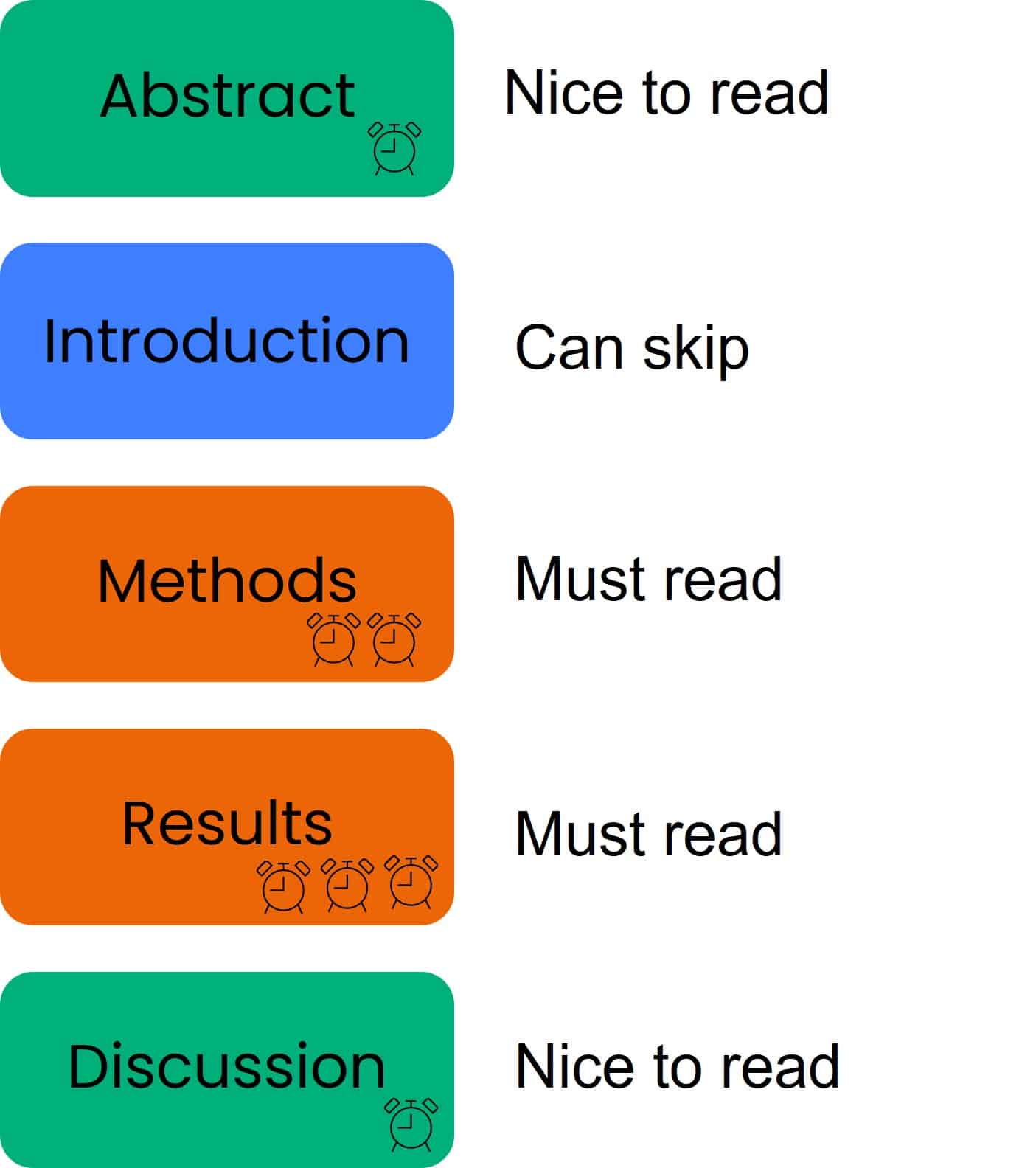
Step 1: Read the Title and Abstract
The title and abstract will give you an overview of the paper’s key points. Most importantly, it will indicate if you should continue and read the rest of the paper. The abstract is often able to view before purchasing or downloading an article, so it can save time and money to read this before committing to the full paper.
Checklist: What to Look for in the Abstract
- The type of journal article. Was it a systematic review? Clinical trial? Meta-analysis?
- The aim. What were they trying to do?
- The experimental setup. Was it in vivo or in vitro, or in silico?
- The key results. What did they find?
- The author’s conclusions. What does it mean? How does it impact the wider field?
Step 2: Skip the Introduction
The introduction is mostly background, and if you are already familiar with the literature, you can scan through or skip this as you probably know it all anyway. You can always return to the introduction if you have time after reading the meatier parts of the paper.
Checklist: What to Look for in the Introduction
- Is the cited literature up to date?
- Do the authors cite only review articles or primary research articles?
- Do they miss key papers?
Step 3: Scan the Methods
Don’t get too bogged down in the methods unless you are researching a new product or technique. Unless the paper details a particularly novel method, just scan through. However, don’t completely ignore the methods section, as the methods used will help you determine the validity of the results.
You should aim to match the methods with the results to understand what has been done. This should be done when reviewing the figures rather than reading the methods section in isolation.
A Note about qPCR Data
If the data is qPCR, take the time to look even more carefully at the methods. According to the MIQE guidelines , the authors need to explain the nucleic acid purification method, yields, and purities, which kits they used, how they determined the efficiency of their assays, and how many replicates they did. There are a lot of factors that can influence qPCR data, and if the paper is leaving out some of the information, you can’t make accurate conclusions from the data.
Checklist: What to Look for in the Methods Section
- Are the controls described? Are they appropriate?
- Are the methods the right choice for the aims of the experiment?
- Did they modify commercial kits, and if so, do they explain how?
- Do they cite previous work to explain methods? If so, access and read the original article to ensure what has been done.
- Ensure adherence to relevant guidelines, e.g., MIQE guidelines for qPCR data.
Step 4: Focus on the Figures
If you want to read a scientific paper effectively, the results section is where you should spend most of your time. This is because the results are the meat of the paper, without which the paper has no purpose.
How you “read” the results is important because while the text is good to read, it is just a description of the results by the author. The author may say that the protein expression levels changed significantly, but you need to look at the results and confirm the change really was significant.
While we hope that authors don’t exaggerate their results, it can be easy to manipulate figures to make them seem more astonishing than they are. We’d also hope this sort of thing would be picked up during editorial and peer review, but peer review can be a flawed process !
Don’t forget any supplementary figures and tables. Just because they are supplementary doesn’t mean they aren’t important. Some of the most important (but not exciting) results are often found here.
We’re not advocating you avoid reading the text of the results section; you certainly should. Just don’t take the authors’ word as gospel. The saying “a picture speaks a thousand words” really is true. Your job is to make sure they match what the author is saying.
And as we mentioned above, read the methods alongside the results and match the method to each figure and table, so you are sure what was done.
A Note About Figure Manipulation
Unfortunately, figure manipulation can be a problem in scientific articles, and while the peer-review process should detect instances of inappropriate manipulation, sometimes things are missed.
And what do we mean about inappropriate manipulation? Not all figure and image manipulation is wrong. Sometimes a western blot needs more brightness or contrast to see the results clearly. This is fine if it is applied to the whole image, but not if it is selectively applied to particular areas. Sometimes there is real intent to deceive, with cases of images swapped, cropped, touched up, or repeated.
Graphs are particularly susceptible to image manipulation, with alterations to graphs changing how the data appears and a reader’s interpretation of a graph. Not starting the axis at 0 can make small differences appear bigger, or vice versa if a scale is too large on the axis. So make sure you pay careful attention to graphs and check the axes (yes, that’s the plural of axis) are appropriate (Figure 1). You should also check if graphs have error bars, and if so, what are they, and is that appropriate?
Statistics can scare many biologists, but it’s important to look at the statistical test and determine if the method is appropriate for the data. Also, be wary of blindly following p-values . You may find situations when an author says something is significant because the statistical test shows a significant p-value, but you can see from the data that it doesn’t look significant. Statistics are not infallible and can be fairly easily manipulated .
Checklist: What to Look for When Reviewing Results
- Are there appropriate scales on graphs?
- Do they use valid statistical analysis? Are results really significant?
- Have they used sufficient n numbers?
- Are the controls appropriate? Should additional controls have been used?
- Is the methodology clear and appropriate?
- Have any figures been inappropriately manipulated?
- Check the supplementary results and methods.
Step 5: Tackle the discussion
The discussion is a great place to determine if you’ve understood the results and the overall message of the paper. It is worth spending more time on the discussion than the introduction as it molds the paper’s results into a story and helps you visualize where they fit in with the overall picture. You should again be wary of authors overinflating their work’s importance and use your judgment to determine if their assertions about what they’ve shown match yours.
One good way to summarize the results of a paper and show how they fit with the wider literature is to sketch out the overall conclusions and how it fits with the current landscape. For example, if the article talks about a specific signaling pathway step, sketch out the pathway with the findings from the paper included. This can help to see the bigger picture, highlight, ensure you understand the impact of the paper, and highlight any unanswered questions.
Test Yourself
A useful exercise when learning how to read a scientific paper (when you have the time!) is to black out the abstract, read the paper and then write an abstract. Then compare the paper’s abstract to the one you wrote. This will demonstrate whether or not you are picking up the paper’s most important point and take-home message.
Checklist: What to Look For in the Discussion Section
- Do you agree with the author’s interpretation of their results?
- Do the results fit with the wider literature?
- Are the authors being objective?
- Do the authors comment on relevant literature and discuss discrepancies between their data and the wider literature?
- Are there any unanswered questions?
Step 6: File it Away
Spending a little time filing your read papers away now can save you A LOT of time in the future (e.g., when writing your own papers or thesis). Use a reference management system and ensure that the entry includes:
- the full and correct citation;
- a very brief summary of the article’s key methods and results;
- any comments or concerns you have;
- any appropriate tags.
Ways to Sharpen Your Critical Analysis Skills
While this article should get you off to a good start, like any muscle, your critical analysis skills need regular workouts to get bigger and better. But how can you hone these skills?
Attend Journal Clubs
Your critical thinking skills benefit dramatically from outside input. This is why journal clubs are so valuable. If your department runs a regular journal club, make sure you attend. If they don’t, set one up. Hearing the views of others can help hone your own critical thinking and allow you to see things from other perspectives. For help and advice on preparing and presenting a journal club session, read our ultimate guide to journal clubs .
Read Online Reviews
Whether in the comments section of the article published online, on a preprint server, or on sites such as PubPeer and Retraction Watch , spend time digesting the views of others. But make sure you apply the same critical analysis skill to these comments and reviews.
These sites can be a useful tool to highlight errors or manipulation you may have missed, but taking these reviews and comments at face value is just as problematic as taking the author’s conclusions as truth. What biases might these reviews have that affect their view? Do you agree with what they say and why?
Final Thoughts on How to Read A Scientific Paper
Reading a scientific paper requires a methodical approach and a critical (but not negative) mindset to ensure that you fully understand what the paper shows.
Reading a paper can seem daunting, and it can be time-consuming if you go in unprepared. However, the process is quicker and smoother once you know how to approach a paper, including what you can and can’t skim. If you don’t have enough time, you can still read a paper effectively without reading the entire paper. Figure 2 highlights what sections can be skimmed and which sections need more of your attention.
Another tip for being more productive (and it’s better for the environment) is to read your papers on-screen . It’ll save time scrambling through a stack of papers and manually filing them away.
Do you have any tips on how to read a scientific paper? Let us know in the comments below.
Want an on-hand checklist to help you analyze papers efficiently despite being busy with research? Download our free article summary and checklist template.
For more tips on keeping track of the scientific literature, head to the Bitesize Bio Managing the Scientific Literature Hub .
Originally published November 20, 2013. Updated and revised September 2022.
Methods can often be important, to judge whether to even trust the results!
The most important is to save all articles that possibly can be interesting in your reference managing system, and classify them with a relevant tag, so that they can be easily found later. Many articles you don’t realize how important they might be until later on. Then you’ll need to find that article you only read the abstract of six months earlier.
Forgot your password?
Lost your password? Please enter your email address. You will receive mail with link to set new password.
Back to login

How to Read Scientific Papers & Research Articles Effectively

Ever tried to read a scientific paper? If you’re finding it difficult, you aren’t alone! Many people struggle to understand scientific literature. No matter your education or background, learning how to read science is a skill that takes time to develop.
Even though it is a challenge, scientific articles contain valuable information. When fully understood, that information can help people make decisions crucial to their lives.
Anyone—even non-scientists—can learn how to read science articles. So grab a warm drink and get comfortable. We’ve written a research reading guide with eight steps to make reading academic papers easy!

Download PDF: How to Be Your Own Research Detective
Why You Should Learn How to Read Research Articles
Peer-reviewed articles were once trusted sources for the latest breaking and noteworthy discoveries in the world of science and medicine. Today, many fields of study are influenced and controlled by companies and entities that have little interest in educating and informing the public.
Many journals and scientific papers have devolved to become a form of advertising for products and questionable methods.
It is up to you, the consumer, to read and decide if something is the truth or if someone is twisting data to influence the public and ultimately sell more products.
Here’s the unfortunate reality: just because it is published in a scientific journal does not mean it is fact. For that reason, the ability to detect the truth while reading a paper is invaluable.
Don’t allow yourself to be fooled! Today, we’ll show you how to critically read a scientific paper. Check out the full guide: How to Become a Research Detective . Or keep reading to learn eight science reading tips you can rely on.
One more thing before we get started – big thanks to Aaron Hertz , the research detective who inspired us and helped to create this guide. Check his satirical yet informative guide to cooking data and stay for his incredible investigative work !
8 Steps: How to Read a Research Paper
If you want to learn how to read a scientific article, you’ll find a lot of the same advice. Most guides will tell you that skimming the abstract, introduction, methods, and results section will be enough to gain a “basic understanding” of whatever you are reading.
However, to fully understand, you’ll need to know more than the basic anatomy of science writing. Instead, you need advice that teaches you what to look for and how to think .
- Always Read the Disclosure Section
- Check the Published Date of the Paper
- Skim All the Sections of the Paper
- Read the Introduction
- Identify How the Paper Fits into the Field of Research
- Read the Discussion Section
- Read the Abstract
- Read the Methods and Results Section
1. Always Read the Disclosure Section
This section is crucial to decipher whether the study is biased. The disclosures section will reveal whether the study was conducted independently or whether a person, company or other group had an impact on the study outcome. A study should ideally not have any conflicts of interest.
If the section shows that the researchers have received money from a company or work for a university that is receiving money from a drug company, they are not independent researchers. You should stop here and dismiss the study.
If you are unsure or if another entity is sponsoring the research, find out who is involved in the noted organizations and see if they have another agenda or receive the support of companies.
This requires a little time and detective work. Do you see that they have received support from any companies? Do the researchers have investments in the company’s drug? Are they receiving monies from an organization that supports a company?

2. Check the Published Date of the Paper
Is this research up to date?
Knowing the publication date will help you determine whether these are the most recent findings. Sometimes additional research has been done since a study’s publication date.
One way investigators can manipulate data is by releasing some data first to create a certain belief and then quietly releasing the rest later. This is an effective strategy for manipulating public opinion.
3. Skim All the Sections of the Paper
Make notes for yourself while reading each section to help evaluate the study and clarify questions you may have. As you go along, take notes, and look up the definitions of any words you’re unsure of.
If you come across an acronym later in a work, a helpful suggestion is to use “CTRL F” on the keyboard to search for the first time it is mentioned, as here is where it will be defined.
Note the definitions, the sample population, the method of testing, and other figures and tables, and/or important facts that can impact the study outcome.
4. Read the Introduction
Read the introduction carefully to learn more about the background of the subject. This includes past research on the subject and the factors that led the researchers to choose to conduct this study. If you are not familiar with the subject, take your time to learn more about it.
As you learn more about the subject, you should also check out some of the references in the introduction.

5. Identify How the Paper Fits into the Field of Research
Does this paper fit in with the field of research and with the special topics of interest or have the authors tampered with the data? What is the principal issue this paper is attempting to address? Will you be better able to comprehend the work’s significance and motivation after reading and analyzing the paper?
What is the researcher’s rationale for studying this intervention or drug? Are there safe alternatives available? Is there a financial incentive for the researchers to draw a particular conclusion?
6. Read the Discussion Section
The discussion section is where you find the paper’s data findings.
The discussion section of the paper is where the data findings are explained and the “story unfolds about the subject matter”. In this section, the samples and measuring tools are presented. The effectiveness of the study is discussed along with whether the study confirmed or disproved the hypothesis. Unfortunately, here the narrative can also be controlled.
7. Read the Abstract
Here is where you find the general summary of the material.
The study’s main objectives, the method of investigation, the key findings, the overview of the interpretations, and the conclusions are often summarized in the abstract. Compare the abstract’s important points to the information offered in the paper’s other sections, such as the discussion, the results, and the conclusions sections.
It will be important to consider the Methods section when looking at the abstract to check to see if the abstract reflects what the data is showing in the conclusion of the study.
8. Read the Methods and Results Section
These are the most complex sections of the study and often where data can be most manipulated.
When reading the results and methods sections, it’s crucial to keep the following things in mind:
- Sample size
- Statistical significance
- Graphics and tables — do they match the conclusions?
- Supplemental materials
Aaron Hertz spends a lot of time in his Substack explaining the numerous ways researchers can manipulate and cherry-pick data to slant the outcome of the study in their favor.
Wrapping up
Navigating the world of scientific research isn’t reserved for those with PhDs or lab coats. Anyone can become adept at understanding these studies with patience, practice, and the right tools. Remember, questioning the source and the motives behind the study is as essential as understanding its content.
Hopefully, these eight steps will act as your compass in the vast sea of research papers. For a more in-depth understanding and additional tips, refer to our full PDF guide: “How to Become a Research Detective.” Knowledge is power, and it’s time to empower yourself!
- Return to my search
Go to Tools & Guides
More on: Guide | how to read research | how to read research papers | Infographic | Research | research papers | Scientific Papers

We are a 100% donor-supported 501(c)(3) non-profit organization. Our work would not be possible without you! Help us expand our reach and share life-saving research.
Mailing Address
FLCCC Alliance 2001 L St. NW Suite 500 Washington, DC 20036
[email protected]
Clinical Support
About the FLCCC
Testimonials
Latest Updates
Newsletter Signup
About FLCCC
©2020–2023 All Rights Reserved FLCCC Alliance. The information contained or presented on this website is for educational purposes only. Information on this site is NOT intended to serve as a substitute for diagnosis, treatment, or advice from a qualified, licensed medical professional. The facts presented are offered as information only in order to empower you – our protocol is not medical advice – and in no way should anyone infer that we, even though we are physicians, or anyone appearing in any content on this website are practicing medicine, it is for educational purposes only. Any treatment protocol you undertake should be discussed with your physician or other licensed medical professional. Seek the advice of a medical professional for proper application of ANY material on this site or our program to your specific situation. NEVER stop or change your medications without consulting your physician. If you are having an emergency contact your emergency services: in the USA that’s 911. FLCCC does not use SMS to conduct marketing campaigns, however, we are required to provide the following disclaimer: By providing my phone number to “FLCCC”, I agree and acknowledge that “FLCCC” may send text messages to my wireless phone number for any purpose. Message and data rates may apply. Message frequency will vary, and you will be able to Opt-out by replying “STOP”. For more information on how your data will be handled please see our privacy policy . Please read our complete disclaimers .
© Copyright 2023. FLCCC Alliance. All Rights Reserved.
Terms and Conditions | Privacy Policy
How to read a scientific research paper
Affiliation.
- 1 Department of Anesthesiology, University of Virginia Health System, PO Box 800710, Charlottesville, VA 22908-0170, USA. [email protected]
- PMID: 19796417
Reading is the most common way that adults learn. With the exponential growth in information, no one has time to read all they need. Reading original research, although difficult, is rewarding and important for growth. Building on past knowledge, the reader should select papers about which he already holds an opinion. Rather than starting at the beginning, this author suggests approaching a paper by reading the conclusions in the abstract first. The methods should be next reviewed, then the results--first in the abstract, and then the full paper. For efficiency, at each step, reasons should be sought not to read any further in the paper. By using this approach, new knowledge will be obtained and many papers will be evaluated, read, and considered.
- Education, Medical, Continuing*
- Information Dissemination
- Peer Review, Research*
- Periodicals as Topic*
- Research Design

An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- PMC10802960

How, and why, science and health researchers read scientific (IMRAD) papers
Frances shiely.
1 Trials Research and Methodologies Unit, HRB Clinical Research Facility, University College Cork, Cork, Ireland
2 School of Public Health, University College Cork, Cork, Ireland
Kerrie Gallagher
Seán r. millar, associated data.
The datasets generated and/or analysed during the current study are available https://osf.io/up4ny/ .
The purpose of our study was to determine the order in which science and health researchers read scientific papers, their reasons for doing so and the perceived difficulty and perceived importance of each section.
Study design and setting
An online survey open to science and health academics and researchers distributed via existing research networks, X (formerly Twitter), and LinkedIn.
Almost 90% of respondents self-declared to be experienced in reading research papers. 98.6% of the sample read the abstract first because it provides an overview of the paper and facilitates a decision on continuing to read on or not. Seventy-five percent perceived it to be the easiest to read and 62.4% perceived it to be very important (highest rank on a 5-point Likert scale). The majority of respondents did not read a paper in the IMRAD (Introduction, Methods, Results And Discussion) format. Perceived difficulty and perceived importance influenced reading order.
Science and health researchers do not typically read scientific and health research papers in IMRAD format. The more important a respondent perceives a section to be, the more likely they are to read it. The easier a section is perceived, the more likely it will be read. We present recommendations to those teaching the skill of writing scientific papers and reports.
Introduction
Reporting in the form of a peer-reviewed research paper, also known as a journal publication or research manuscript, is essential to the healthcare and science professions. The skill of writing a peer reviewed paper is highly specialized and challenging. It is also a challenge to teach this skill, yet it is essential to do so, as students are often required to engage with complex academic texts as well as write scientific reports [ 1 – 3 ]. Other cited reasons are: (i) increasing scientific literacy; (ii) staying informed of progress in a particular field of study; (iii) understanding the causation, clinical features, and natural history of a disease; (iv) evaluating the effectiveness of diagnostic tests and clinical therapies; and (v) determining whether there is support for or opposition to a particular argument [ 4 – 6 ]. Additionally, it is imperative that the reader is able to identify robustly designed research in order to make informed recommendations regarding policy or patient care [ 7 ].
Currently, most healthcare research papers are presented in the IMRAD format: I ntroduction (why the authors decided to do the research), M ethods (how they did it and how they chose to analyse their results), R esults (what they found), A nd D iscussion (what they believe the results to mean) with a preceding abstract. However, there is no evidence-based research determining the suitability of this approach. Nevertheless, it provides a means for scientific communities to organise and structure their work effectively [ 8 ]. Within the scientific community, the approach is based on the notion that having a clear structure and procedures can help scientists produce better quality work. In addition, it is thought to reduce the risk of mistakes and oversights and ensure compliance with best practices in research (10). We know that the amount of time students spend reading academic material is estimated to be between seven and fourteen hours per week, which represents an important component of the academic schedule [ 3 , 9 , 10 ]. Therefore, research on the suitability of the IMRAD approach is important.
Professor Trisha Greenhalgh, author of the seminal text “How to read a paper: the basics of evidence-based medicine and healthcare” suggests that if you are deciding whether a paper is worthy of study, you should do so based on the design of the methods section [ 11 ]. This is largely opinion based and is not predicated on clear evidence. Anecdotal evidence suggests people choose to read and examine research papers in different ways, but the literature is scant on the topic. One UK study attempts to address the strategies used by researchers and students when reading primary research [ 12 ]. The authors report that individuals at different career stages value different sections of scientific papers, with novice readers finding the methods and results sections to be particularly challenging to decipher [ 12 ]. Similarly, a study conducted in the US examined and compared how faculty members and students in an undergraduate science course engaged with a primary research article [ 13 ]. Faculty and students were able to demonstrate understanding of the research design at some point during the reading process, however, the faculty displayed this ability almost four times as often as students [ 13 ]. Both of these studies are limited in their capacity and generalisability as they are restricted to students and researchers in the biological sciences.
From a teaching and learning perspective, we are interested in knowing more about how science and health researchers read IMRAD research papers and the importance they place on each section. Our primary aim is to establish the order in which these researchers read an IMRAD formatted paper and why. By establishing this, educators can better craft their teaching to ensure that students understand the importance of each section, have the knowledge and skills necessary to write an effective scientific paper or report, and the ability to critically appraise the work of others.
Materials and methods
The survey ( S1 File ) was created on Google Forms by two members of the research team (KG and FS) and independently reviewed by two reviewers (ST and EM). The survey had three parts. Part 1 was concerned with written informed consent. When participants clicked the link, they were brought the informed consent page which provided details of the study, what was required from them, knowledge of the voluntary nature of the participation and right to withdraw at any stage, and the contact details of the principal investigator. To proceed, participants had to select either I consent to participate, which brought the participants to Part 2 of the survey, or I do not consent to participate, which meant the participants exited the survey. Part 2 of the survey collected data concerning the demographic characteristics of the respondents. Part 3 focused on questions pertaining to the order in which researchers read a primary research paper, how easy it is to read each of the sections (based on a 7-point scale) and how important each section of a primary research paper is for its understanding. The survey also assessed when a reader stops reading and why. The style of questions was mixed and included Likert scale ratings and closed and open-ended questions. Ethical approval was granted by the Social Research Ethics Committee (SREC), University College Cork (Log 2021–165). Participants provided written informed consent.
Recruitment
This was an online survey and recruitment was online. Inclusion criteria were: academics, health professionals, and patients and members of the public, involved in science or health research and/or teaching. Exclusion criterion was: under 18 years of age. Our recruitment strategy was to target academics, researchers and patient and public involvement members of our existing networks, all who work within or are affiliated with Universities in the UK, Ireland and Canada. The lead author, FS, is primarily associated with clinical trial networks. An email of invitation outlining the aim of the study and survey link was sent electronically via the Health Research Trial Methodology Research Network (HRB TMRN), Ireland (~3000 subscribers), Medial Research Council-National Institutes of Health and Care Research-Trial Methodology Research Partnership (MRC-NIHR-TMRP), UK, locally at University, University College Cork (all academic and research staff—~2500 people), via X, formerly known as Twitter (@FrancesShiely; @hrbtmrn) which was forwarded and liked and via LinkedIn (FS account). FS also distributed the link to her academic research partners in Ireland, UK, Hungary, Czech Republic, France, and Canada and asked them to forward to their respective Universities and contacts.
Statistical analysis
We obtained 152 responses to the survey, 139 of which completed answers to the order in which they read the research paper. These were included in the analyses. Descriptive characteristics were examined for the full sample. Likert scale answers to reading order, perceived difficulty and perceived importance questions for each research paper section are shown as percentages. Reading order, perceived difficulty and importance ranking were also examined according to career stage. Observations were independent, with no individuals belonging to more than one career stage group. Relationships between perceived difficulty ranking, perceived importance ranking and research paper reading order were also examined using Spearman’s rank-order correlation. Data analysis was conducted using Stata SE Version 13 (Stata Corporation, College Station, TX, USA) for Windows. For all analyses, a p value (two-tailed) of less than .05 was considered to indicate statistical significance. Qualitative variables were summarised according to the most frequent occurrence to provide a picture on the reasons participants chose to read a paper in their chosen format.
Table 1 shows descriptive characteristics of the study respondents. The majority of subjects were female (61.2%), 90.7% were under 60 years of age and 94.7% reside in Europe. Study respondents included MSc and PhD students (n = 17), early-career researchers (n = 39), mid-career researchers (n = 36) and established/leading researchers and research managers (n = 39). A majority (88.5%) worked in academic research at a university or college, with 61.9% indicating both research and teaching responsibilities. Almost 90% of respondents stated that they were experienced in reading a research paper.
Reading order, perceived difficulty and perceived importance for IRMAD sections
Fig 1 shows research paper reading order according to each section. A majority of respondents (98.6%) indicated that they read the abstract section first when reading a scientific paper, with 36.0% (Introduction), 29.5% (Methods), 36.0% (Results–text), 31.7% (Results–figures & tables), 43.9% (Discussion) and 36.7% (Conclusion) of subjects stating that they read these sections second, third, fourth, fifth, sixth, and last, respectively. Noticeably, a majority of respondents indicated that they did not read a paper in the IMRAD order; for instance, while over one-third of respondents stated that they read the Introduction section second, 64.0% did not, with just over one-fifth (20.9%) indicating that they read this section last.

We asked respondents why they read in their preferred order. For the 98.6% (149/152) who selected the abstract first, the reasons can be summarised as the fact the abstract gives an overview or summary of the paper and it allows one to see if the paper is worth continuing reading (“the abstract gives the summary and informs as to whether I will read the whole paper”, “abstract gives a feel for quality of the paper”, “fast and easy”, “the abstract has the main points and is usually freely available”). We were interested in what respondents read after the abstract. For those who read the introduction second, i.e., the IMRAD format (only two read the introduction first), the dominant reason was because it’s logical to read in the order it’s presented (“it’s the logical order”, “I usually read papers in the order it is written”). For those who chose the methods second, 21.2% (31/146) the reasons can be themed as to ensure robustness or quality of the study (“methods to understand whether it was well conducted”, “checking the methods to ensure it is relevant to me”, “understand how the methods led to such results”, “is this something I can trust”). Only 15 people read the results section second, regardless of whether it was the results-text or results-figures & tables. The key reasons for reading the results second can be summarised as establishing the findings (“get right to the results”, “results to understand the main findings”, “the results are arguably the most important part of the document”, “do the results show what they are saying?”). Those choosing to read the discussion second, 8.1% (12/149), did so to establish the key findings (“the discussion and conclusion is the essence of what the study found”, “discussion and conclusion are most interesting”, “discussion to see if anything interesting came out of the results”). For those who read the conclusion second, 17% (25/146), the reasons are summarised as establishing the overall view of the paper and if the research is of value “see if a paper of value”, “know if it’s useful to me”, “final result/outcome”, “overall view”, “clarifies what the author perceives to have been achieved”, “I read the conclusions to build on the summary conclusions of the abstract”.
To explore perceived difficulty when reading a research paper, participants were asked to rank a series of questions according to reading difficulty on a 7-point scale, 1 being the easiest and 7 being the most difficult ( Fig 2 ). Similar to reading order, a majority (75.0%) of respondents stated that they found the Abstract section to be easiest to read (rank 1). The Introduction and Conclusion sections were perceived as next easiest, respectively. Taking the blue and orange together (ranks 1 and 2) the same trend applies. On the opposite end of the scale (rank 7-most difficult, the dark navy colour), the Results-figures & tables section, was perceived to be most difficult (26.3%), followed by the Methods (25.6%) and Results-text (17.3%).

Perceived importance for each section was assessed using a 5-point Likert scale ( Fig 3 ). The Abstract and Methods sections were perceived as very important by 62.4% and 58.8% of respondents, respectively. Although few respondents perceived any section as unimportant or very unimportant, only 29.5%, 31.8% and 32.6% of subjects believed that the Introduction, Discussion and Conclusion sections were very important.

Reading order and career stage
Fig 4 shows the different sections of the research paper in reading order according to career stage. Differences in reading order were noted, with the greatest differences in reading order observed in the Results-text, Results-figures & tables and Discussion sections. Notably, 46.2% of established/leading researchers or research managers read the Results-text section fourth (in IMRAD order), compared to 29.4% of MSc by research/PhD students and 28.2% of mid-career researchers who did so. Similarly, a higher percentage of established/leading researchers or research managers indicated reading the Results-figures & tables and Discussion sections according to IMRAD reading order when compared to other career stages.

Perceived difficulty and importance for each section according to career stage
Perceived difficulty ranking and perceived importance ranking according to career stage, for each IMRAD section and the abstract, are shown in Figs Figs5 5 and and6. 6 . Consistent with results observed among all subjects, regardless of career stage, the Results-text and Results-figures & tables sections and Discussion sections were perceived as most difficult to read. Differences were found to be greatest for the Conclusion section, with MSc by research or PhD students being more likely to rank this section as difficult to read. With regard to the Introduction section, mid-career researchers were more likely to rank this section as important. Interestingly, MSc by research/PhD students were more likely to rank the Methods section as being important when compared to other career stages.

Correlations between perceived difficulty, importance and reading order
Spearman correlation coefficients between the ranking of perceived difficulty, perceived importance and reading order, according to each section, are shown in Table 2 . Significant correlations between perceived difficulty ranking and reading order were observed for the Methods (rho = 0.450, p < .001), Results-figures & tables (rho = 0.333, p < .001), Discussion (rho = 0.204, p = .018) and Conclusion (rho = 0.334, p < .001) sections, indicating that the easier a respondent perceived that section to read, the more likely they were to read it at an earlier stage. Significant correlations between perceived importance ranking and reading order were observed for the Introduction (rho = 0.467, p < .001), Methods (rho = 0.426, p < .001), Results (text) (rho = 0.250, p = .003), Results (figures & tables) (rho = 0.173, p = .048), Discussion (rho = 0.214, p = 0.14) and Conclusion (rho = 0.302, p < .001) sections, suggesting that the more important a respondent perceived that section to be, the more likely they were to read it at an earlier stage.
Values are presented as Spearman correlation coefficients (rho) between perceived difficulty ranking, perceived importance ranking and research paper reading order, for each section (n = 139).
Significant p highlighted .
When and why respondents stop reading a paper
We asked respondents why they stopped reading a research paper. The main sections were (in no particular order) results, introduction, methods and abstract. Of the 98.6% that read the abstract first, 28% (42/149) then stopped reading at this stage. The reason given in all cases is lack of relevance (“not relevant to my interests”, “will have identified if it is of relevance”). The main reasons for those that stop reading at the introduction is the writing style (“poorly written”, if the writing style is overly complex”, it’s too dense and not interesting”, not relevant, poorly conducted”. For those who stop reading at the results section, the main reasons given are the results are too complex or poorly explained (“it gets too difficult to understand”, “paper is not relevant”, “too complicated”, “it’s no longer relevant if results are not clear”). For those who stop reading at the methods section, the main reason is they are unclear or too difficult or there is a perception that the methods are not needed (“becomes technical and I don’t’ need more details”, the methods might not be interesting to what I am trying to learn from reading the paper”, generally for my work methods are not important”, not interested in it, I Know already by the methods if I ‘like’ the paper”, “most often the methodology is not clear enough”).
We know most research papers are published in IMRAD format, preceded by an abstract. We sought to establish if researchers, and at different career stages, typically read a paper in this way. We found that even though most researchers consider themselves experienced readers of primary research papers, respondents did not typically engage with the literature in IMRAD format. Reading strategies varied depending on perceived difficulty and perceived importance of the paper sections. The more important a respondent perceived the section to be, the more likely they were to read it at an earlier stage. Almost all science and health researchers read the abstract first, and a significant proportion stop reading there. The primary reason for stopping is lack of relevance.
We can see very clearly that the abstract is read first by most researchers, regardless of career stage, perceived to be the most important, and also perceived to be the easiest to read. While there isn’t prior research to compare this finding to, we surmise that it is because it is a summary of the overall paper, and a logical place to begin. It’s also possible that it is because it is presented first in all research papers, or for pay per view journals/papers it is usually available when the rest of a paper is beyond a paywall. It could be that researchers know the abstract is used by journal editors to decide if it is worth continuing to read a paper to decide if it should be peer-reviewed or it could be that the abstract is often used in systematic reviews when screening (typically title and abstract) and thus habitually researchers read it first. However, we don’t know any of this for sure and would need a qualitative study to confirm or refute these. We do know the reasons that a significant number of people stop reading the abstract and that is relevance, or the lack of it. Another consideration might be the reading level at which the abstract is pitched. We have conducted a previous study on the readability of trial lay summaries, which are written for a lay audience [ 14 ]. We found that no lay summary met the recommended reading age for health care information of 11–12 years. None of them were considered "easy" to read, in fact over 85% were considered "difficult" to read [ 14 ]. By extension we can assume the scientific abstract, which we have considered here, will be no better, and likely worse, and thus challenging for a novice researcher. We recommend that when teaching the skill of writing a scientific paper, or report, in the science or health research disciplines, teachers emphasise the importance of the abstract and give consideration to the target audience when formulating their approach. Online readability tools are readily available to assist the process, but we do not recommend relying on them solely [ 14 ].
Further evidence of the complexity of reading different parts of scientific papers is the response to the methods section. The evidence that does exist on how to read a paper suggests that if you are deciding whether a paper is worthy of study, you should do so based on the design of the methods section. As mentioned previously, this was opinion based rather than evidence based [ 3 ]. Our respondents typically considered the methods section to be of low importance on the 7-point Likert Scale and some stopped reading there due to the unclear language or technical nature of the section. This result was unexpected, as there is a general consensus in the scientific community that the methods section is considered one of the most important sections of any research paper, given that it provides essential insight into the conduct of the study and its integrity, the conclusions derived from them, and the reproducibility of the work [ 3 ]. Indeed, across several well recognized and validated critical appraisal tools, including the Critical Appraisal Skills Program (CASP) for Randomized Control Trials [ 15 ], the ROB 2.0 Risk of Bias Tool [ 16 ] and the ROBINS-I Risk of Bias for non-randomized (observational) studies [ 17 ], focus is directed towards systematically examining the methods section in order for the reader to determine the strength of the evidence presented, its reliability and relevance to clinical practice.
Strengths and weaknesses
We had a reasonable response (n = 139) but we are unable to calculate a response rate due to the mode of distribution, a significant weakness to the study. Our study likely demonstrates selection bias, given the known research networks through which the survey was distributed are all funded through academic grants and the majority of our respondents were academic researchers working in a University/College. The research would be enhanced if we had a larger response from non-profit organisations. There were only six respondents outside of Europe and this is a weakness in terms of generalisability. However, on the positive side, non-response bias was not evident, and we had a full dataset for 139 respondents.
Recommendations
The lessons for future practice are:
- Ensure your abstract gives enough detail to ensure relevance and pique interest because if you don’t, science and health researchers will lose interest and stop reading;
- Ensure the introduction is well written because if it is poorly written, you will lose the reader (we suggest using the freely available online readability scales, e.g., Flesch Reading Ease Score (FRES), Flesch-Kincaid Grade Level (FKGL), Simplified Measure of Gobbledegook (SMOG), Gunning Fog (GF), Coleman-Liau Index (CLI), and Automated Readability Index (ARI) readability scales and paying attention to Plain Language guidelines [ 18 ], e.g., in the UK the Plain English UK guidelines are most relevant;
- Don’t make the methods section too technical. Find the balance between overcomplicating the methods and giving enough detail so the study can be replicated;
- Keep the results simple and explain them well.
Conclusions
This study provides an insight into the order in which IMRAD papers are read and the reasons for doing so. Existing evidence says that to determine if a paper is worthy of reading, you should read the methods section to decide. Our results refute this and show the methods section to be one of the sections perceived most difficult to read and also the least important. Our results show the importance of the abstract to the scientific and health research community, and we recommend when teaching the skill of scientific writing a particular focus is given to the abstract. Future research on this topic is welcome in a more diverse and larger sample.
Supporting information
Acknowledgments.
We would like to thank those who took the time to fill in our questionnaire.
Funding Statement
This study was funded by Teaching and Learning Enhancement Fund at University College Cork for a summer student scholarship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
Loading metrics
Open Access
Ten simple rules for reading a scientific paper
* E-mail: [email protected]
Affiliation Division of Infectious Diseases and International Health, Department of Medicine, University of Virginia School of Medicine, Charlottesville, Virginia, United States of America
- Maureen A. Carey,
- Kevin L. Steiner,
- William A. Petri Jr

Published: July 30, 2020
- https://doi.org/10.1371/journal.pcbi.1008032
- Reader Comments
Citation: Carey MA, Steiner KL, Petri WA Jr (2020) Ten simple rules for reading a scientific paper. PLoS Comput Biol 16(7): e1008032. https://doi.org/10.1371/journal.pcbi.1008032
Editor: Scott Markel, Dassault Systemes BIOVIA, UNITED STATES
Copyright: © 2020 Carey et al. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Funding: MAC was supported by the PhRMA Foundation's Postdoctoral Fellowship in Translational Medicine and Therapeutics and the University of Virginia's Engineering-in-Medicine seed grant, and KLS was supported by the NIH T32 Global Biothreats Training Program at the University of Virginia (AI055432). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests: The authors have declared that no competing interests exist.
Introduction
“There is no problem that a library card can't solve” according to author Eleanor Brown [ 1 ]. This advice is sound, probably for both life and science, but even the best tool (like the library) is most effective when accompanied by instructions and a basic understanding of how and when to use it.
For many budding scientists, the first day in a new lab setting often involves a stack of papers, an email full of links to pertinent articles, or some promise of a richer understanding so long as one reads enough of the scientific literature. However, the purpose and approach to reading a scientific article is unlike that of reading a news story, novel, or even a textbook and can initially seem unapproachable. Having good habits for reading scientific literature is key to setting oneself up for success, identifying new research questions, and filling in the gaps in one’s current understanding; developing these good habits is the first crucial step.
Advice typically centers around two main tips: read actively and read often. However, active reading, or reading with an intent to understand, is both a learned skill and a level of effort. Although there is no one best way to do this, we present 10 simple rules, relevant to novices and seasoned scientists alike, to teach our strategy for active reading based on our experience as readers and as mentors of undergraduate and graduate researchers, medical students, fellows, and early career faculty. Rules 1–5 are big picture recommendations. Rules 6–8 relate to philosophy of reading. Rules 9–10 guide the “now what?” questions one should ask after reading and how to integrate what was learned into one’s own science.
Rule 1: Pick your reading goal
What you want to get out of an article should influence your approach to reading it. Table 1 includes a handful of example intentions and how you might prioritize different parts of the same article differently based on your goals as a reader.
- PPT PowerPoint slide
- PNG larger image
- TIFF original image
https://doi.org/10.1371/journal.pcbi.1008032.t001
Rule 2: Understand the author’s goal
In written communication, the reader and the writer are equally important. Both influence the final outcome: in this case, your scientific understanding! After identifying your goal, think about the author’s goal for sharing this project. This will help you interpret the data and understand the author’s interpretation of the data. However, this requires some understanding of who the author(s) are (e.g., what are their scientific interests?), the scientific field in which they work (e.g., what techniques are available in this field?), and how this paper fits into the author’s research (e.g., is this work building on an author’s longstanding project or controversial idea?). This information may be hard to glean without experience and a history of reading. But don’t let this be a discouragement to starting the process; it is by the act of reading that this experience is gained!
A good step toward understanding the goal of the author(s) is to ask yourself: What kind of article is this? Journals publish different types of articles, including methods, review, commentary, resources, and research articles as well as other types that are specific to a particular journal or groups of journals. These article types have different formatting requirements and expectations for content. Knowing the article type will help guide your evaluation of the information presented. Is the article a methods paper, presenting a new technique? Is the article a review article, intended to summarize a field or problem? Is it a commentary, intended to take a stand on a controversy or give a big picture perspective on a problem? Is it a resource article, presenting a new tool or data set for others to use? Is it a research article, written to present new data and the authors’ interpretation of those data? The type of paper, and its intended purpose, will get you on your way to understanding the author’s goal.
Rule 3: Ask six questions
When reading, ask yourself: (1) What do the author(s) want to know (motivation)? (2) What did they do (approach/methods)? (3) Why was it done that way (context within the field)? (4) What do the results show (figures and data tables)? (5) How did the author(s) interpret the results (interpretation/discussion)? (6) What should be done next? (Regarding this last question, the author(s) may provide some suggestions in the discussion, but the key is to ask yourself what you think should come next.)
Each of these questions can and should be asked about the complete work as well as each table, figure, or experiment within the paper. Early on, it can take a long time to read one article front to back, and this can be intimidating. Break down your understanding of each section of the work with these questions to make the effort more manageable.
Rule 4: Unpack each figure and table
Scientists write original research papers primarily to present new data that may change or reinforce the collective knowledge of a field. Therefore, the most important parts of this type of scientific paper are the data. Some people like to scrutinize the figures and tables (including legends) before reading any of the “main text”: because all of the important information should be obtained through the data. Others prefer to read through the results section while sequentially examining the figures and tables as they are addressed in the text. There is no correct or incorrect approach: Try both to see what works best for you. The key is making sure that one understands the presented data and how it was obtained.
For each figure, work to understand each x- and y-axes, color scheme, statistical approach (if one was used), and why the particular plotting approach was used. For each table, identify what experimental groups and variables are presented. Identify what is shown and how the data were collected. This is typically summarized in the legend or caption but often requires digging deeper into the methods: Do not be afraid to refer back to the methods section frequently to ensure a full understanding of how the presented data were obtained. Again, ask the questions in Rule 3 for each figure or panel and conclude with articulating the “take home” message.
Rule 5: Understand the formatting intentions
Just like the overall intent of the article (discussed in Rule 2), the intent of each section within a research article can guide your interpretation. Some sections are intended to be written as objective descriptions of the data (i.e., the Results section), whereas other sections are intended to present the author’s interpretation of the data. Remember though that even “objective” sections are written by and, therefore, influenced by the authors interpretations. Check out Table 2 to understand the intent of each section of a research article. When reading a specific paper, you can also refer to the journal’s website to understand the formatting intentions. The “For Authors” section of a website will have some nitty gritty information that is less relevant for the reader (like word counts) but will also summarize what the journal editors expect in each section. This will help to familiarize you with the goal of each article section.
https://doi.org/10.1371/journal.pcbi.1008032.t002
Rule 6: Be critical
Published papers are not truths etched in stone. Published papers in high impact journals are not truths etched in stone. Published papers by bigwigs in the field are not truths etched in stone. Published papers that seem to agree with your own hypothesis or data are not etched in stone. Published papers that seem to refute your hypothesis or data are not etched in stone.
Science is a never-ending work in progress, and it is essential that the reader pushes back against the author’s interpretation to test the strength of their conclusions. Everyone has their own perspective and may interpret the same data in different ways. Mistakes are sometimes published, but more often these apparent errors are due to other factors such as limitations of a methodology and other limits to generalizability (selection bias, unaddressed, or unappreciated confounders). When reading a paper, it is important to consider if these factors are pertinent.
Critical thinking is a tough skill to learn but ultimately boils down to evaluating data while minimizing biases. Ask yourself: Are there other, equally likely, explanations for what is observed? In addition to paying close attention to potential biases of the study or author(s), a reader should also be alert to one’s own preceding perspective (and biases). Take time to ask oneself: Do I find this paper compelling because it affirms something I already think (or wish) is true? Or am I discounting their findings because it differs from what I expect or from my own work?
The phenomenon of a self-fulfilling prophecy, or expectancy, is well studied in the psychology literature [ 2 ] and is why many studies are conducted in a “blinded” manner [ 3 ]. It refers to the idea that a person may assume something to be true and their resultant behavior aligns to make it true. In other words, as humans and scientists, we often find exactly what we are looking for. A scientist may only test their hypotheses and fail to evaluate alternative hypotheses; perhaps, a scientist may not be aware of alternative, less biased ways to test her or his hypothesis that are typically used in different fields. Individuals with different life, academic, and work experiences may think of several alternative hypotheses, all equally supported by the data.
Rule 7: Be kind
The author(s) are human too. So, whenever possible, give them the benefit of the doubt. An author may write a phrase differently than you would, forcing you to reread the sentence to understand it. Someone in your field may neglect to cite your paper because of a reference count limit. A figure panel may be misreferenced as Supplemental Fig 3E when it is obviously Supplemental Fig 4E. While these things may be frustrating, none are an indication that the quality of work is poor. Try to avoid letting these minor things influence your evaluation and interpretation of the work.
Similarly, if you intend to share your critique with others, be extra kind. An author (especially the lead author) may invest years of their time into a single paper. Hearing a kindly phrased critique can be difficult but constructive. Hearing a rude, brusque, or mean-spirited critique can be heartbreaking, especially for young scientists or those seeking to establish their place within a field and who may worry that they do not belong.
Rule 8: Be ready to go the extra mile
To truly understand a scientific work, you often will need to look up a term, dig into the supplemental materials, or read one or more of the cited references. This process takes time. Some advisors recommend reading an article three times: The first time, simply read without the pressure of understanding or critiquing the work. For the second time, aim to understand the paper. For the third read through, take notes.
Some people engage with a paper by printing it out and writing all over it. The reader might write question marks in the margins to mark parts (s)he wants to return to, circle unfamiliar terms (and then actually look them up!), highlight or underline important statements, and draw arrows linking figures and the corresponding interpretation in the discussion. Not everyone needs a paper copy to engage in the reading process but, whatever your version of “printing it out” is, do it.

Rule 9: Talk about it
Talking about an article in a journal club or more informal environment forces active reading and participation with the material. Studies show that teaching is one of the best ways to learn and that teachers learn the material even better as the teaching task becomes more complex [ 4 – 5 ]; anecdotally, such observations inspired the phrase “to teach is to learn twice.”
Beyond formal settings such as journal clubs, lab meetings, and academic classes, discuss papers with your peers, mentors, and colleagues in person or electronically. Twitter and other social media platforms have become excellent resources for discussing papers with other scientists, the public or your nonscientist friends, or even the paper’s author(s). Describing a paper can be done at multiple levels and your description can contain all of the scientific details, only the big picture summary, or perhaps the implications for the average person in your community. All of these descriptions will solidify your understanding, while highlighting gaps in your knowledge and informing those around you.
Rule 10: Build on it
One approach we like to use for communicating how we build on the scientific literature is by starting research presentations with an image depicting a wall of Lego bricks. Each brick is labeled with the reference for a paper, and the wall highlights the body of literature on which the work is built. We describe the work and conclusions of each paper represented by a labeled brick and discuss each brick and the wall as a whole. The top brick on the wall is left blank: We aspire to build on this work and label this brick with our own work. We then delve into our own research, discoveries, and the conclusions it inspires. We finish our presentations with the image of the Legos and summarize our presentation on that empty brick.
Whether you are reading an article to understand a new topic area or to move a research project forward, effective learning requires that you integrate knowledge from multiple sources (“click” those Lego bricks together) and build upwards. Leveraging published work will enable you to build a stronger and taller structure. The first row of bricks is more stable once a second row is assembled on top of it and so on and so forth. Moreover, the Lego construction will become taller and larger if you build upon the work of others, rather than using only your own bricks.
Build on the article you read by thinking about how it connects to ideas described in other papers and within own work, implementing a technique in your own research, or attempting to challenge or support the hypothesis of the author(s) with a more extensive literature review. Integrate the techniques and scientific conclusions learned from an article into your own research or perspective in the classroom or research lab. You may find that this process strengthens your understanding, leads you toward new and unexpected interests or research questions, or returns you back to the original article with new questions and critiques of the work. All of these experiences are part of the “active reading”: process and are signs of a successful reading experience.
In summary, practice these rules to learn how to read a scientific article, keeping in mind that this process will get easier (and faster) with experience. We are firm believers that an hour in the library will save a week at the bench; this diligent practice will ultimately make you both a more knowledgeable and productive scientist. As you develop the skills to read an article, try to also foster good reading and learning habits for yourself (recommendations here: [ 6 ] and [ 7 ], respectively) and in others. Good luck and happy reading!
Acknowledgments
Thank you to the mentors, teachers, and students who have shaped our thoughts on reading, learning, and what science is all about.
- 1. Brown E. The Weird Sisters. G. P. Putnam’s Sons; 2011.
- View Article
- Google Scholar
- PubMed/NCBI
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 26 March 2024
Predicting and improving complex beer flavor through machine learning
- Michiel Schreurs ORCID: orcid.org/0000-0002-9449-5619 1 , 2 , 3 na1 ,
- Supinya Piampongsant 1 , 2 , 3 na1 ,
- Miguel Roncoroni ORCID: orcid.org/0000-0001-7461-1427 1 , 2 , 3 na1 ,
- Lloyd Cool ORCID: orcid.org/0000-0001-9936-3124 1 , 2 , 3 , 4 ,
- Beatriz Herrera-Malaver ORCID: orcid.org/0000-0002-5096-9974 1 , 2 , 3 ,
- Christophe Vanderaa ORCID: orcid.org/0000-0001-7443-5427 4 ,
- Florian A. Theßeling 1 , 2 , 3 ,
- Łukasz Kreft ORCID: orcid.org/0000-0001-7620-4657 5 ,
- Alexander Botzki ORCID: orcid.org/0000-0001-6691-4233 5 ,
- Philippe Malcorps 6 ,
- Luk Daenen 6 ,
- Tom Wenseleers ORCID: orcid.org/0000-0002-1434-861X 4 &
- Kevin J. Verstrepen ORCID: orcid.org/0000-0002-3077-6219 1 , 2 , 3
Nature Communications volume 15 , Article number: 2368 ( 2024 ) Cite this article
51k Accesses
856 Altmetric
Metrics details
- Chemical engineering
- Gas chromatography
- Machine learning
- Metabolomics
- Taste receptors
The perception and appreciation of food flavor depends on many interacting chemical compounds and external factors, and therefore proves challenging to understand and predict. Here, we combine extensive chemical and sensory analyses of 250 different beers to train machine learning models that allow predicting flavor and consumer appreciation. For each beer, we measure over 200 chemical properties, perform quantitative descriptive sensory analysis with a trained tasting panel and map data from over 180,000 consumer reviews to train 10 different machine learning models. The best-performing algorithm, Gradient Boosting, yields models that significantly outperform predictions based on conventional statistics and accurately predict complex food features and consumer appreciation from chemical profiles. Model dissection allows identifying specific and unexpected compounds as drivers of beer flavor and appreciation. Adding these compounds results in variants of commercial alcoholic and non-alcoholic beers with improved consumer appreciation. Together, our study reveals how big data and machine learning uncover complex links between food chemistry, flavor and consumer perception, and lays the foundation to develop novel, tailored foods with superior flavors.
Similar content being viewed by others
BitterSweet: Building machine learning models for predicting the bitter and sweet taste of small molecules
Rudraksh Tuwani, Somin Wadhwa & Ganesh Bagler
Sensory lexicon and aroma volatiles analysis of brewing malt
Xiaoxia Su, Miao Yu, … Tianyi Du
Predicting odor from molecular structure: a multi-label classification approach
Kushagra Saini & Venkatnarayan Ramanathan
Introduction
Predicting and understanding food perception and appreciation is one of the major challenges in food science. Accurate modeling of food flavor and appreciation could yield important opportunities for both producers and consumers, including quality control, product fingerprinting, counterfeit detection, spoilage detection, and the development of new products and product combinations (food pairing) 1 , 2 , 3 , 4 , 5 , 6 . Accurate models for flavor and consumer appreciation would contribute greatly to our scientific understanding of how humans perceive and appreciate flavor. Moreover, accurate predictive models would also facilitate and standardize existing food assessment methods and could supplement or replace assessments by trained and consumer tasting panels, which are variable, expensive and time-consuming 7 , 8 , 9 . Lastly, apart from providing objective, quantitative, accurate and contextual information that can help producers, models can also guide consumers in understanding their personal preferences 10 .
Despite the myriad of applications, predicting food flavor and appreciation from its chemical properties remains a largely elusive goal in sensory science, especially for complex food and beverages 11 , 12 . A key obstacle is the immense number of flavor-active chemicals underlying food flavor. Flavor compounds can vary widely in chemical structure and concentration, making them technically challenging and labor-intensive to quantify, even in the face of innovations in metabolomics, such as non-targeted metabolic fingerprinting 13 , 14 . Moreover, sensory analysis is perhaps even more complicated. Flavor perception is highly complex, resulting from hundreds of different molecules interacting at the physiochemical and sensorial level. Sensory perception is often non-linear, characterized by complex and concentration-dependent synergistic and antagonistic effects 15 , 16 , 17 , 18 , 19 , 20 , 21 that are further convoluted by the genetics, environment, culture and psychology of consumers 22 , 23 , 24 . Perceived flavor is therefore difficult to measure, with problems of sensitivity, accuracy, and reproducibility that can only be resolved by gathering sufficiently large datasets 25 . Trained tasting panels are considered the prime source of quality sensory data, but require meticulous training, are low throughput and high cost. Public databases containing consumer reviews of food products could provide a valuable alternative, especially for studying appreciation scores, which do not require formal training 25 . Public databases offer the advantage of amassing large amounts of data, increasing the statistical power to identify potential drivers of appreciation. However, public datasets suffer from biases, including a bias in the volunteers that contribute to the database, as well as confounding factors such as price, cult status and psychological conformity towards previous ratings of the product.
Classical multivariate statistics and machine learning methods have been used to predict flavor of specific compounds by, for example, linking structural properties of a compound to its potential biological activities or linking concentrations of specific compounds to sensory profiles 1 , 26 . Importantly, most previous studies focused on predicting organoleptic properties of single compounds (often based on their chemical structure) 27 , 28 , 29 , 30 , 31 , 32 , 33 , thus ignoring the fact that these compounds are present in a complex matrix in food or beverages and excluding complex interactions between compounds. Moreover, the classical statistics commonly used in sensory science 34 , 35 , 36 , 37 , 38 , 39 require a large sample size and sufficient variance amongst predictors to create accurate models. They are not fit for studying an extensive set of hundreds of interacting flavor compounds, since they are sensitive to outliers, have a high tendency to overfit and are less suited for non-linear and discontinuous relationships 40 .
In this study, we combine extensive chemical analyses and sensory data of a set of different commercial beers with machine learning approaches to develop models that predict taste, smell, mouthfeel and appreciation from compound concentrations. Beer is particularly suited to model the relationship between chemistry, flavor and appreciation. First, beer is a complex product, consisting of thousands of flavor compounds that partake in complex sensory interactions 41 , 42 , 43 . This chemical diversity arises from the raw materials (malt, yeast, hops, water and spices) and biochemical conversions during the brewing process (kilning, mashing, boiling, fermentation, maturation and aging) 44 , 45 . Second, the advent of the internet saw beer consumers embrace online review platforms, such as RateBeer (ZX Ventures, Anheuser-Busch InBev SA/NV) and BeerAdvocate (Next Glass, inc.). In this way, the beer community provides massive data sets of beer flavor and appreciation scores, creating extraordinarily large sensory databases to complement the analyses of our professional sensory panel. Specifically, we characterize over 200 chemical properties of 250 commercial beers, spread across 22 beer styles, and link these to the descriptive sensory profiling data of a 16-person in-house trained tasting panel and data acquired from over 180,000 public consumer reviews. These unique and extensive datasets enable us to train a suite of machine learning models to predict flavor and appreciation from a beer’s chemical profile. Dissection of the best-performing models allows us to pinpoint specific compounds as potential drivers of beer flavor and appreciation. Follow-up experiments confirm the importance of these compounds and ultimately allow us to significantly improve the flavor and appreciation of selected commercial beers. Together, our study represents a significant step towards understanding complex flavors and reinforces the value of machine learning to develop and refine complex foods. In this way, it represents a stepping stone for further computer-aided food engineering applications 46 .
To generate a comprehensive dataset on beer flavor, we selected 250 commercial Belgian beers across 22 different beer styles (Supplementary Fig. S1 ). Beers with ≤ 4.2% alcohol by volume (ABV) were classified as non-alcoholic and low-alcoholic. Blonds and Tripels constitute a significant portion of the dataset (12.4% and 11.2%, respectively) reflecting their presence on the Belgian beer market and the heterogeneity of beers within these styles. By contrast, lager beers are less diverse and dominated by a handful of brands. Rare styles such as Brut or Faro make up only a small fraction of the dataset (2% and 1%, respectively) because fewer of these beers are produced and because they are dominated by distinct characteristics in terms of flavor and chemical composition.
Extensive analysis identifies relationships between chemical compounds in beer
For each beer, we measured 226 different chemical properties, including common brewing parameters such as alcohol content, iso-alpha acids, pH, sugar concentration 47 , and over 200 flavor compounds (Methods, Supplementary Table S1 ). A large portion (37.2%) are terpenoids arising from hopping, responsible for herbal and fruity flavors 16 , 48 . A second major category are yeast metabolites, such as esters and alcohols, that result in fruity and solvent notes 48 , 49 , 50 . Other measured compounds are primarily derived from malt, or other microbes such as non- Saccharomyces yeasts and bacteria (‘wild flora’). Compounds that arise from spices or staling are labeled under ‘Others’. Five attributes (caloric value, total acids and total ester, hop aroma and sulfur compounds) are calculated from multiple individually measured compounds.
As a first step in identifying relationships between chemical properties, we determined correlations between the concentrations of the compounds (Fig. 1 , upper panel, Supplementary Data 1 and 2 , and Supplementary Fig. S2 . For the sake of clarity, only a subset of the measured compounds is shown in Fig. 1 ). Compounds of the same origin typically show a positive correlation, while absence of correlation hints at parameters varying independently. For example, the hop aroma compounds citronellol, and alpha-terpineol show moderate correlations with each other (Spearman’s rho=0.39 and 0.57), but not with the bittering hop component iso-alpha acids (Spearman’s rho=0.16 and −0.07). This illustrates how brewers can independently modify hop aroma and bitterness by selecting hop varieties and dosage time. If hops are added early in the boiling phase, chemical conversions increase bitterness while aromas evaporate, conversely, late addition of hops preserves aroma but limits bitterness 51 . Similarly, hop-derived iso-alpha acids show a strong anti-correlation with lactic acid and acetic acid, likely reflecting growth inhibition of lactic acid and acetic acid bacteria, or the consequent use of fewer hops in sour beer styles, such as West Flanders ales and Fruit beers, that rely on these bacteria for their distinct flavors 52 . Finally, yeast-derived esters (ethyl acetate, ethyl decanoate, ethyl hexanoate, ethyl octanoate) and alcohols (ethanol, isoamyl alcohol, isobutanol, and glycerol), correlate with Spearman coefficients above 0.5, suggesting that these secondary metabolites are correlated with the yeast genetic background and/or fermentation parameters and may be difficult to influence individually, although the choice of yeast strain may offer some control 53 .

Spearman rank correlations are shown. Descriptors are grouped according to their origin (malt (blue), hops (green), yeast (red), wild flora (yellow), Others (black)), and sensory aspect (aroma, taste, palate, and overall appreciation). Please note that for the chemical compounds, for the sake of clarity, only a subset of the total number of measured compounds is shown, with an emphasis on the key compounds for each source. For more details, see the main text and Methods section. Chemical data can be found in Supplementary Data 1 , correlations between all chemical compounds are depicted in Supplementary Fig. S2 and correlation values can be found in Supplementary Data 2 . See Supplementary Data 4 for sensory panel assessments and Supplementary Data 5 for correlation values between all sensory descriptors.
Interestingly, different beer styles show distinct patterns for some flavor compounds (Supplementary Fig. S3 ). These observations agree with expectations for key beer styles, and serve as a control for our measurements. For instance, Stouts generally show high values for color (darker), while hoppy beers contain elevated levels of iso-alpha acids, compounds associated with bitter hop taste. Acetic and lactic acid are not prevalent in most beers, with notable exceptions such as Kriek, Lambic, Faro, West Flanders ales and Flanders Old Brown, which use acid-producing bacteria ( Lactobacillus and Pediococcus ) or unconventional yeast ( Brettanomyces ) 54 , 55 . Glycerol, ethanol and esters show similar distributions across all beer styles, reflecting their common origin as products of yeast metabolism during fermentation 45 , 53 . Finally, low/no-alcohol beers contain low concentrations of glycerol and esters. This is in line with the production process for most of the low/no-alcohol beers in our dataset, which are produced through limiting fermentation or by stripping away alcohol via evaporation or dialysis, with both methods having the unintended side-effect of reducing the amount of flavor compounds in the final beer 56 , 57 .
Besides expected associations, our data also reveals less trivial associations between beer styles and specific parameters. For example, geraniol and citronellol, two monoterpenoids responsible for citrus, floral and rose flavors and characteristic of Citra hops, are found in relatively high amounts in Christmas, Saison, and Brett/co-fermented beers, where they may originate from terpenoid-rich spices such as coriander seeds instead of hops 58 .
Tasting panel assessments reveal sensorial relationships in beer
To assess the sensory profile of each beer, a trained tasting panel evaluated each of the 250 beers for 50 sensory attributes, including different hop, malt and yeast flavors, off-flavors and spices. Panelists used a tasting sheet (Supplementary Data 3 ) to score the different attributes. Panel consistency was evaluated by repeating 12 samples across different sessions and performing ANOVA. In 95% of cases no significant difference was found across sessions ( p > 0.05), indicating good panel consistency (Supplementary Table S2 ).
Aroma and taste perception reported by the trained panel are often linked (Fig. 1 , bottom left panel and Supplementary Data 4 and 5 ), with high correlations between hops aroma and taste (Spearman’s rho=0.83). Bitter taste was found to correlate with hop aroma and taste in general (Spearman’s rho=0.80 and 0.69), and particularly with “grassy” noble hops (Spearman’s rho=0.75). Barnyard flavor, most often associated with sour beers, is identified together with stale hops (Spearman’s rho=0.97) that are used in these beers. Lactic and acetic acid, which often co-occur, are correlated (Spearman’s rho=0.66). Interestingly, sweetness and bitterness are anti-correlated (Spearman’s rho = −0.48), confirming the hypothesis that they mask each other 59 , 60 . Beer body is highly correlated with alcohol (Spearman’s rho = 0.79), and overall appreciation is found to correlate with multiple aspects that describe beer mouthfeel (alcohol, carbonation; Spearman’s rho= 0.32, 0.39), as well as with hop and ester aroma intensity (Spearman’s rho=0.39 and 0.35).
Similar to the chemical analyses, sensorial analyses confirmed typical features of specific beer styles (Supplementary Fig. S4 ). For example, sour beers (Faro, Flanders Old Brown, Fruit beer, Kriek, Lambic, West Flanders ale) were rated acidic, with flavors of both acetic and lactic acid. Hoppy beers were found to be bitter and showed hop-associated aromas like citrus and tropical fruit. Malt taste is most detected among scotch, stout/porters, and strong ales, while low/no-alcohol beers, which often have a reputation for being ‘worty’ (reminiscent of unfermented, sweet malt extract) appear in the middle. Unsurprisingly, hop aromas are most strongly detected among hoppy beers. Like its chemical counterpart (Supplementary Fig. S3 ), acidity shows a right-skewed distribution, with the most acidic beers being Krieks, Lambics, and West Flanders ales.
Tasting panel assessments of specific flavors correlate with chemical composition
We find that the concentrations of several chemical compounds strongly correlate with specific aroma or taste, as evaluated by the tasting panel (Fig. 2 , Supplementary Fig. S5 , Supplementary Data 6 ). In some cases, these correlations confirm expectations and serve as a useful control for data quality. For example, iso-alpha acids, the bittering compounds in hops, strongly correlate with bitterness (Spearman’s rho=0.68), while ethanol and glycerol correlate with tasters’ perceptions of alcohol and body, the mouthfeel sensation of fullness (Spearman’s rho=0.82/0.62 and 0.72/0.57 respectively) and darker color from roasted malts is a good indication of malt perception (Spearman’s rho=0.54).

Heatmap colors indicate Spearman’s Rho. Axes are organized according to sensory categories (aroma, taste, mouthfeel, overall), chemical categories and chemical sources in beer (malt (blue), hops (green), yeast (red), wild flora (yellow), Others (black)). See Supplementary Data 6 for all correlation values.
Interestingly, for some relationships between chemical compounds and perceived flavor, correlations are weaker than expected. For example, the rose-smelling phenethyl acetate only weakly correlates with floral aroma. This hints at more complex relationships and interactions between compounds and suggests a need for a more complex model than simple correlations. Lastly, we uncovered unexpected correlations. For instance, the esters ethyl decanoate and ethyl octanoate appear to correlate slightly with hop perception and bitterness, possibly due to their fruity flavor. Iron is anti-correlated with hop aromas and bitterness, most likely because it is also anti-correlated with iso-alpha acids. This could be a sign of metal chelation of hop acids 61 , given that our analyses measure unbound hop acids and total iron content, or could result from the higher iron content in dark and Fruit beers, which typically have less hoppy and bitter flavors 62 .
Public consumer reviews complement expert panel data
To complement and expand the sensory data of our trained tasting panel, we collected 180,000 reviews of our 250 beers from the online consumer review platform RateBeer. This provided numerical scores for beer appearance, aroma, taste, palate, overall quality as well as the average overall score.
Public datasets are known to suffer from biases, such as price, cult status and psychological conformity towards previous ratings of a product. For example, prices correlate with appreciation scores for these online consumer reviews (rho=0.49, Supplementary Fig. S6 ), but not for our trained tasting panel (rho=0.19). This suggests that prices affect consumer appreciation, which has been reported in wine 63 , while blind tastings are unaffected. Moreover, we observe that some beer styles, like lagers and non-alcoholic beers, generally receive lower scores, reflecting that online reviewers are mostly beer aficionados with a preference for specialty beers over lager beers. In general, we find a modest correlation between our trained panel’s overall appreciation score and the online consumer appreciation scores (Fig. 3 , rho=0.29). Apart from the aforementioned biases in the online datasets, serving temperature, sample freshness and surroundings, which are all tightly controlled during the tasting panel sessions, can vary tremendously across online consumers and can further contribute to (among others, appreciation) differences between the two categories of tasters. Importantly, in contrast to the overall appreciation scores, for many sensory aspects the results from the professional panel correlated well with results obtained from RateBeer reviews. Correlations were highest for features that are relatively easy to recognize even for untrained tasters, like bitterness, sweetness, alcohol and malt aroma (Fig. 3 and below).
RateBeer text mining results can be found in Supplementary Data 7 . Rho values shown are Spearman correlation values, with asterisks indicating significant correlations ( p < 0.05, two-sided). All p values were smaller than 0.001, except for Esters aroma (0.0553), Esters taste (0.3275), Esters aroma—banana (0.0019), Coriander (0.0508) and Diacetyl (0.0134).
Besides collecting consumer appreciation from these online reviews, we developed automated text analysis tools to gather additional data from review texts (Supplementary Data 7 ). Processing review texts on the RateBeer database yielded comparable results to the scores given by the trained panel for many common sensory aspects, including acidity, bitterness, sweetness, alcohol, malt, and hop tastes (Fig. 3 ). This is in line with what would be expected, since these attributes require less training for accurate assessment and are less influenced by environmental factors such as temperature, serving glass and odors in the environment. Consumer reviews also correlate well with our trained panel for 4-vinyl guaiacol, a compound associated with a very characteristic aroma. By contrast, correlations for more specific aromas like ester, coriander or diacetyl are underrepresented in the online reviews, underscoring the importance of using a trained tasting panel and standardized tasting sheets with explicit factors to be scored for evaluating specific aspects of a beer. Taken together, our results suggest that public reviews are trustworthy for some, but not all, flavor features and can complement or substitute taste panel data for these sensory aspects.
Models can predict beer sensory profiles from chemical data
The rich datasets of chemical analyses, tasting panel assessments and public reviews gathered in the first part of this study provided us with a unique opportunity to develop predictive models that link chemical data to sensorial features. Given the complexity of beer flavor, basic statistical tools such as correlations or linear regression may not always be the most suitable for making accurate predictions. Instead, we applied different machine learning models that can model both simple linear and complex interactive relationships. Specifically, we constructed a set of regression models to predict (a) trained panel scores for beer flavor and quality and (b) public reviews’ appreciation scores from beer chemical profiles. We trained and tested 10 different models (Methods), 3 linear regression-based models (simple linear regression with first-order interactions (LR), lasso regression with first-order interactions (Lasso), partial least squares regressor (PLSR)), 5 decision tree models (AdaBoost regressor (ABR), extra trees (ET), gradient boosting regressor (GBR), random forest (RF) and XGBoost regressor (XGBR)), 1 support vector regression (SVR), and 1 artificial neural network (ANN) model.
To compare the performance of our machine learning models, the dataset was randomly split into a training and test set, stratified by beer style. After a model was trained on data in the training set, its performance was evaluated on its ability to predict the test dataset obtained from multi-output models (based on the coefficient of determination, see Methods). Additionally, individual-attribute models were ranked per descriptor and the average rank was calculated, as proposed by Korneva et al. 64 . Importantly, both ways of evaluating the models’ performance agreed in general. Performance of the different models varied (Table 1 ). It should be noted that all models perform better at predicting RateBeer results than results from our trained tasting panel. One reason could be that sensory data is inherently variable, and this variability is averaged out with the large number of public reviews from RateBeer. Additionally, all tree-based models perform better at predicting taste than aroma. Linear models (LR) performed particularly poorly, with negative R 2 values, due to severe overfitting (training set R 2 = 1). Overfitting is a common issue in linear models with many parameters and limited samples, especially with interaction terms further amplifying the number of parameters. L1 regularization (Lasso) successfully overcomes this overfitting, out-competing multiple tree-based models on the RateBeer dataset. Similarly, the dimensionality reduction of PLSR avoids overfitting and improves performance, to some extent. Still, tree-based models (ABR, ET, GBR, RF and XGBR) show the best performance, out-competing the linear models (LR, Lasso, PLSR) commonly used in sensory science 65 .
GBR models showed the best overall performance in predicting sensory responses from chemical information, with R 2 values up to 0.75 depending on the predicted sensory feature (Supplementary Table S4 ). The GBR models predict consumer appreciation (RateBeer) better than our trained panel’s appreciation (R 2 value of 0.67 compared to R 2 value of 0.09) (Supplementary Table S3 and Supplementary Table S4 ). ANN models showed intermediate performance, likely because neural networks typically perform best with larger datasets 66 . The SVR shows intermediate performance, mostly due to the weak predictions of specific attributes that lower the overall performance (Supplementary Table S4 ).
Model dissection identifies specific, unexpected compounds as drivers of consumer appreciation
Next, we leveraged our models to infer important contributors to sensory perception and consumer appreciation. Consumer preference is a crucial sensory aspects, because a product that shows low consumer appreciation scores often does not succeed commercially 25 . Additionally, the requirement for a large number of representative evaluators makes consumer trials one of the more costly and time-consuming aspects of product development. Hence, a model for predicting chemical drivers of overall appreciation would be a welcome addition to the available toolbox for food development and optimization.
Since GBR models on our RateBeer dataset showed the best overall performance, we focused on these models. Specifically, we used two approaches to identify important contributors. First, rankings of the most important predictors for each sensorial trait in the GBR models were obtained based on impurity-based feature importance (mean decrease in impurity). High-ranked parameters were hypothesized to be either the true causal chemical properties underlying the trait, to correlate with the actual causal properties, or to take part in sensory interactions affecting the trait 67 (Fig. 4A ). In a second approach, we used SHAP 68 to determine which parameters contributed most to the model for making predictions of consumer appreciation (Fig. 4B ). SHAP calculates parameter contributions to model predictions on a per-sample basis, which can be aggregated into an importance score.
A The impurity-based feature importance (mean deviance in impurity, MDI) calculated from the Gradient Boosting Regression (GBR) model predicting RateBeer appreciation scores. The top 15 highest ranked chemical properties are shown. B SHAP summary plot for the top 15 parameters contributing to our GBR model. Each point on the graph represents a sample from our dataset. The color represents the concentration of that parameter, with bluer colors representing low values and redder colors representing higher values. Greater absolute values on the horizontal axis indicate a higher impact of the parameter on the prediction of the model. C Spearman correlations between the 15 most important chemical properties and consumer overall appreciation. Numbers indicate the Spearman Rho correlation coefficient, and the rank of this correlation compared to all other correlations. The top 15 important compounds were determined using SHAP (panel B).
Both approaches identified ethyl acetate as the most predictive parameter for beer appreciation (Fig. 4 ). Ethyl acetate is the most abundant ester in beer with a typical ‘fruity’, ‘solvent’ and ‘alcoholic’ flavor, but is often considered less important than other esters like isoamyl acetate. The second most important parameter identified by SHAP is ethanol, the most abundant beer compound after water. Apart from directly contributing to beer flavor and mouthfeel, ethanol drastically influences the physical properties of beer, dictating how easily volatile compounds escape the beer matrix to contribute to beer aroma 69 . Importantly, it should also be noted that the importance of ethanol for appreciation is likely inflated by the very low appreciation scores of non-alcoholic beers (Supplementary Fig. S4 ). Despite not often being considered a driver of beer appreciation, protein level also ranks highly in both approaches, possibly due to its effect on mouthfeel and body 70 . Lactic acid, which contributes to the tart taste of sour beers, is the fourth most important parameter identified by SHAP, possibly due to the generally high appreciation of sour beers in our dataset.
Interestingly, some of the most important predictive parameters for our model are not well-established as beer flavors or are even commonly regarded as being negative for beer quality. For example, our models identify methanethiol and ethyl phenyl acetate, an ester commonly linked to beer staling 71 , as a key factor contributing to beer appreciation. Although there is no doubt that high concentrations of these compounds are considered unpleasant, the positive effects of modest concentrations are not yet known 72 , 73 .
To compare our approach to conventional statistics, we evaluated how well the 15 most important SHAP-derived parameters correlate with consumer appreciation (Fig. 4C ). Interestingly, only 6 of the properties derived by SHAP rank amongst the top 15 most correlated parameters. For some chemical compounds, the correlations are so low that they would have likely been considered unimportant. For example, lactic acid, the fourth most important parameter, shows a bimodal distribution for appreciation, with sour beers forming a separate cluster, that is missed entirely by the Spearman correlation. Additionally, the correlation plots reveal outliers, emphasizing the need for robust analysis tools. Together, this highlights the need for alternative models, like the Gradient Boosting model, that better grasp the complexity of (beer) flavor.
Finally, to observe the relationships between these chemical properties and their predicted targets, partial dependence plots were constructed for the six most important predictors of consumer appreciation 74 , 75 , 76 (Supplementary Fig. S7 ). One-way partial dependence plots show how a change in concentration affects the predicted appreciation. These plots reveal an important limitation of our models: appreciation predictions remain constant at ever-increasing concentrations. This implies that once a threshold concentration is reached, further increasing the concentration does not affect appreciation. This is false, as it is well-documented that certain compounds become unpleasant at high concentrations, including ethyl acetate (‘nail polish’) 77 and methanethiol (‘sulfury’ and ‘rotten cabbage’) 78 . The inability of our models to grasp that flavor compounds have optimal levels, above which they become negative, is a consequence of working with commercial beer brands where (off-)flavors are rarely too high to negatively impact the product. The two-way partial dependence plots show how changing the concentration of two compounds influences predicted appreciation, visualizing their interactions (Supplementary Fig. S7 ). In our case, the top 5 parameters are dominated by additive or synergistic interactions, with high concentrations for both compounds resulting in the highest predicted appreciation.
To assess the robustness of our best-performing models and model predictions, we performed 100 iterations of the GBR, RF and ET models. In general, all iterations of the models yielded similar performance (Supplementary Fig. S8 ). Moreover, the main predictors (including the top predictors ethanol and ethyl acetate) remained virtually the same, especially for GBR and RF. For the iterations of the ET model, we did observe more variation in the top predictors, which is likely a consequence of the model’s inherent random architecture in combination with co-correlations between certain predictors. However, even in this case, several of the top predictors (ethanol and ethyl acetate) remain unchanged, although their rank in importance changes (Supplementary Fig. S8 ).
Next, we investigated if a combination of RateBeer and trained panel data into one consolidated dataset would lead to stronger models, under the hypothesis that such a model would suffer less from bias in the datasets. A GBR model was trained to predict appreciation on the combined dataset. This model underperformed compared to the RateBeer model, both in the native case and when including a dataset identifier (R 2 = 0.67, 0.26 and 0.42 respectively). For the latter, the dataset identifier is the most important feature (Supplementary Fig. S9 ), while most of the feature importance remains unchanged, with ethyl acetate and ethanol ranking highest, like in the original model trained only on RateBeer data. It seems that the large variation in the panel dataset introduces noise, weakening the models’ performances and reliability. In addition, it seems reasonable to assume that both datasets are fundamentally different, with the panel dataset obtained by blind tastings by a trained professional panel.
Lastly, we evaluated whether beer style identifiers would further enhance the model’s performance. A GBR model was trained with parameters that explicitly encoded the styles of the samples. This did not improve model performance (R2 = 0.66 with style information vs R2 = 0.67). The most important chemical features are consistent with the model trained without style information (eg. ethanol and ethyl acetate), and with the exception of the most preferred (strong ale) and least preferred (low/no-alcohol) styles, none of the styles were among the most important features (Supplementary Fig. S9 , Supplementary Table S5 and S6 ). This is likely due to a combination of style-specific chemical signatures, such as iso-alpha acids and lactic acid, that implicitly convey style information to the original models, as well as the low number of samples belonging to some styles, making it difficult for the model to learn style-specific patterns. Moreover, beer styles are not rigorously defined, with some styles overlapping in features and some beers being misattributed to a specific style, all of which leads to more noise in models that use style parameters.
Model validation
To test if our predictive models give insight into beer appreciation, we set up experiments aimed at improving existing commercial beers. We specifically selected overall appreciation as the trait to be examined because of its complexity and commercial relevance. Beer flavor comprises a complex bouquet rather than single aromas and tastes 53 . Hence, adding a single compound to the extent that a difference is noticeable may lead to an unbalanced, artificial flavor. Therefore, we evaluated the effect of combinations of compounds. Because Blond beers represent the most extensive style in our dataset, we selected a beer from this style as the starting material for these experiments (Beer 64 in Supplementary Data 1 ).
In the first set of experiments, we adjusted the concentrations of compounds that made up the most important predictors of overall appreciation (ethyl acetate, ethanol, lactic acid, ethyl phenyl acetate) together with correlated compounds (ethyl hexanoate, isoamyl acetate, glycerol), bringing them up to 95 th percentile ethanol-normalized concentrations (Methods) within the Blond group (‘Spiked’ concentration in Fig. 5A ). Compared to controls, the spiked beers were found to have significantly improved overall appreciation among trained panelists, with panelist noting increased intensity of ester flavors, sweetness, alcohol, and body fullness (Fig. 5B ). To disentangle the contribution of ethanol to these results, a second experiment was performed without the addition of ethanol. This resulted in a similar outcome, including increased perception of alcohol and overall appreciation.
Adding the top chemical compounds, identified as best predictors of appreciation by our model, into poorly appreciated beers results in increased appreciation from our trained panel. Results of sensory tests between base beers and those spiked with compounds identified as the best predictors by the model. A Blond and Non/Low-alcohol (0.0% ABV) base beers were brought up to 95th-percentile ethanol-normalized concentrations within each style. B For each sensory attribute, tasters indicated the more intense sample and selected the sample they preferred. The numbers above the bars correspond to the p values that indicate significant changes in perceived flavor (two-sided binomial test: alpha 0.05, n = 20 or 13).
In a last experiment, we tested whether using the model’s predictions can boost the appreciation of a non-alcoholic beer (beer 223 in Supplementary Data 1 ). Again, the addition of a mixture of predicted compounds (omitting ethanol, in this case) resulted in a significant increase in appreciation, body, ester flavor and sweetness.
Predicting flavor and consumer appreciation from chemical composition is one of the ultimate goals of sensory science. A reliable, systematic and unbiased way to link chemical profiles to flavor and food appreciation would be a significant asset to the food and beverage industry. Such tools would substantially aid in quality control and recipe development, offer an efficient and cost-effective alternative to pilot studies and consumer trials and would ultimately allow food manufacturers to produce superior, tailor-made products that better meet the demands of specific consumer groups more efficiently.
A limited set of studies have previously tried, to varying degrees of success, to predict beer flavor and beer popularity based on (a limited set of) chemical compounds and flavors 79 , 80 . Current sensitive, high-throughput technologies allow measuring an unprecedented number of chemical compounds and properties in a large set of samples, yielding a dataset that can train models that help close the gaps between chemistry and flavor, even for a complex natural product like beer. To our knowledge, no previous research gathered data at this scale (250 samples, 226 chemical parameters, 50 sensory attributes and 5 consumer scores) to disentangle and validate the chemical aspects driving beer preference using various machine-learning techniques. We find that modern machine learning models outperform conventional statistical tools, such as correlations and linear models, and can successfully predict flavor appreciation from chemical composition. This could be attributed to the natural incorporation of interactions and non-linear or discontinuous effects in machine learning models, which are not easily grasped by the linear model architecture. While linear models and partial least squares regression represent the most widespread statistical approaches in sensory science, in part because they allow interpretation 65 , 81 , 82 , modern machine learning methods allow for building better predictive models while preserving the possibility to dissect and exploit the underlying patterns. Of the 10 different models we trained, tree-based models, such as our best performing GBR, showed the best overall performance in predicting sensory responses from chemical information, outcompeting artificial neural networks. This agrees with previous reports for models trained on tabular data 83 . Our results are in line with the findings of Colantonio et al. who also identified the gradient boosting architecture as performing best at predicting appreciation and flavor (of tomatoes and blueberries, in their specific study) 26 . Importantly, besides our larger experimental scale, we were able to directly confirm our models’ predictions in vivo.
Our study confirms that flavor compound concentration does not always correlate with perception, suggesting complex interactions that are often missed by more conventional statistics and simple models. Specifically, we find that tree-based algorithms may perform best in developing models that link complex food chemistry with aroma. Furthermore, we show that massive datasets of untrained consumer reviews provide a valuable source of data, that can complement or even replace trained tasting panels, especially for appreciation and basic flavors, such as sweetness and bitterness. This holds despite biases that are known to occur in such datasets, such as price or conformity bias. Moreover, GBR models predict taste better than aroma. This is likely because taste (e.g. bitterness) often directly relates to the corresponding chemical measurements (e.g., iso-alpha acids), whereas such a link is less clear for aromas, which often result from the interplay between multiple volatile compounds. We also find that our models are best at predicting acidity and alcohol, likely because there is a direct relation between the measured chemical compounds (acids and ethanol) and the corresponding perceived sensorial attribute (acidity and alcohol), and because even untrained consumers are generally able to recognize these flavors and aromas.
The predictions of our final models, trained on review data, hold even for blind tastings with small groups of trained tasters, as demonstrated by our ability to validate specific compounds as drivers of beer flavor and appreciation. Since adding a single compound to the extent of a noticeable difference may result in an unbalanced flavor profile, we specifically tested our identified key drivers as a combination of compounds. While this approach does not allow us to validate if a particular single compound would affect flavor and/or appreciation, our experiments do show that this combination of compounds increases consumer appreciation.
It is important to stress that, while it represents an important step forward, our approach still has several major limitations. A key weakness of the GBR model architecture is that amongst co-correlating variables, the largest main effect is consistently preferred for model building. As a result, co-correlating variables often have artificially low importance scores, both for impurity and SHAP-based methods, like we observed in the comparison to the more randomized Extra Trees models. This implies that chemicals identified as key drivers of a specific sensory feature by GBR might not be the true causative compounds, but rather co-correlate with the actual causative chemical. For example, the high importance of ethyl acetate could be (partially) attributed to the total ester content, ethanol or ethyl hexanoate (rho=0.77, rho=0.72 and rho=0.68), while ethyl phenylacetate could hide the importance of prenyl isobutyrate and ethyl benzoate (rho=0.77 and rho=0.76). Expanding our GBR model to include beer style as a parameter did not yield additional power or insight. This is likely due to style-specific chemical signatures, such as iso-alpha acids and lactic acid, that implicitly convey style information to the original model, as well as the smaller sample size per style, limiting the power to uncover style-specific patterns. This can be partly attributed to the curse of dimensionality, where the high number of parameters results in the models mainly incorporating single parameter effects, rather than complex interactions such as style-dependent effects 67 . A larger number of samples may overcome some of these limitations and offer more insight into style-specific effects. On the other hand, beer style is not a rigid scientific classification, and beers within one style often differ a lot, which further complicates the analysis of style as a model factor.
Our study is limited to beers from Belgian breweries. Although these beers cover a large portion of the beer styles available globally, some beer styles and consumer patterns may be missing, while other features might be overrepresented. For example, many Belgian ales exhibit yeast-driven flavor profiles, which is reflected in the chemical drivers of appreciation discovered by this study. In future work, expanding the scope to include diverse markets and beer styles could lead to the identification of even more drivers of appreciation and better models for special niche products that were not present in our beer set.
In addition to inherent limitations of GBR models, there are also some limitations associated with studying food aroma. Even if our chemical analyses measured most of the known aroma compounds, the total number of flavor compounds in complex foods like beer is still larger than the subset we were able to measure in this study. For example, hop-derived thiols, that influence flavor at very low concentrations, are notoriously difficult to measure in a high-throughput experiment. Moreover, consumer perception remains subjective and prone to biases that are difficult to avoid. It is also important to stress that the models are still immature and that more extensive datasets will be crucial for developing more complete models in the future. Besides more samples and parameters, our dataset does not include any demographic information about the tasters. Including such data could lead to better models that grasp external factors like age and culture. Another limitation is that our set of beers consists of high-quality end-products and lacks beers that are unfit for sale, which limits the current model in accurately predicting products that are appreciated very badly. Finally, while models could be readily applied in quality control, their use in sensory science and product development is restrained by their inability to discern causal relationships. Given that the models cannot distinguish compounds that genuinely drive consumer perception from those that merely correlate, validation experiments are essential to identify true causative compounds.
Despite the inherent limitations, dissection of our models enabled us to pinpoint specific molecules as potential drivers of beer aroma and consumer appreciation, including compounds that were unexpected and would not have been identified using standard approaches. Important drivers of beer appreciation uncovered by our models include protein levels, ethyl acetate, ethyl phenyl acetate and lactic acid. Currently, many brewers already use lactic acid to acidify their brewing water and ensure optimal pH for enzymatic activity during the mashing process. Our results suggest that adding lactic acid can also improve beer appreciation, although its individual effect remains to be tested. Interestingly, ethanol appears to be unnecessary to improve beer appreciation, both for blond beer and alcohol-free beer. Given the growing consumer interest in alcohol-free beer, with a predicted annual market growth of >7% 84 , it is relevant for brewers to know what compounds can further increase consumer appreciation of these beers. Hence, our model may readily provide avenues to further improve the flavor and consumer appreciation of both alcoholic and non-alcoholic beers, which is generally considered one of the key challenges for future beer production.
Whereas we see a direct implementation of our results for the development of superior alcohol-free beverages and other food products, our study can also serve as a stepping stone for the development of novel alcohol-containing beverages. We want to echo the growing body of scientific evidence for the negative effects of alcohol consumption, both on the individual level by the mutagenic, teratogenic and carcinogenic effects of ethanol 85 , 86 , as well as the burden on society caused by alcohol abuse and addiction. We encourage the use of our results for the production of healthier, tastier products, including novel and improved beverages with lower alcohol contents. Furthermore, we strongly discourage the use of these technologies to improve the appreciation or addictive properties of harmful substances.
The present work demonstrates that despite some important remaining hurdles, combining the latest developments in chemical analyses, sensory analysis and modern machine learning methods offers exciting avenues for food chemistry and engineering. Soon, these tools may provide solutions in quality control and recipe development, as well as new approaches to sensory science and flavor research.
Beer selection
250 commercial Belgian beers were selected to cover the broad diversity of beer styles and corresponding diversity in chemical composition and aroma. See Supplementary Fig. S1 .
Chemical dataset
Sample preparation.
Beers within their expiration date were purchased from commercial retailers. Samples were prepared in biological duplicates at room temperature, unless explicitly stated otherwise. Bottle pressure was measured with a manual pressure device (Steinfurth Mess-Systeme GmbH) and used to calculate CO 2 concentration. The beer was poured through two filter papers (Macherey-Nagel, 500713032 MN 713 ¼) to remove carbon dioxide and prevent spontaneous foaming. Samples were then prepared for measurements by targeted Headspace-Gas Chromatography-Flame Ionization Detector/Flame Photometric Detector (HS-GC-FID/FPD), Headspace-Solid Phase Microextraction-Gas Chromatography-Mass Spectrometry (HS-SPME-GC-MS), colorimetric analysis, enzymatic analysis, Near-Infrared (NIR) analysis, as described in the sections below. The mean values of biological duplicates are reported for each compound.
HS-GC-FID/FPD
HS-GC-FID/FPD (Shimadzu GC 2010 Plus) was used to measure higher alcohols, acetaldehyde, esters, 4-vinyl guaicol, and sulfur compounds. Each measurement comprised 5 ml of sample pipetted into a 20 ml glass vial containing 1.75 g NaCl (VWR, 27810.295). 100 µl of 2-heptanol (Sigma-Aldrich, H3003) (internal standard) solution in ethanol (Fisher Chemical, E/0650DF/C17) was added for a final concentration of 2.44 mg/L. Samples were flushed with nitrogen for 10 s, sealed with a silicone septum, stored at −80 °C and analyzed in batches of 20.
The GC was equipped with a DB-WAXetr column (length, 30 m; internal diameter, 0.32 mm; layer thickness, 0.50 µm; Agilent Technologies, Santa Clara, CA, USA) to the FID and an HP-5 column (length, 30 m; internal diameter, 0.25 mm; layer thickness, 0.25 µm; Agilent Technologies, Santa Clara, CA, USA) to the FPD. N 2 was used as the carrier gas. Samples were incubated for 20 min at 70 °C in the headspace autosampler (Flow rate, 35 cm/s; Injection volume, 1000 µL; Injection mode, split; Combi PAL autosampler, CTC analytics, Switzerland). The injector, FID and FPD temperatures were kept at 250 °C. The GC oven temperature was first held at 50 °C for 5 min and then allowed to rise to 80 °C at a rate of 5 °C/min, followed by a second ramp of 4 °C/min until 200 °C kept for 3 min and a final ramp of (4 °C/min) until 230 °C for 1 min. Results were analyzed with the GCSolution software version 2.4 (Shimadzu, Kyoto, Japan). The GC was calibrated with a 5% EtOH solution (VWR International) containing the volatiles under study (Supplementary Table S7 ).
HS-SPME-GC-MS
HS-SPME-GC-MS (Shimadzu GCMS-QP-2010 Ultra) was used to measure additional volatile compounds, mainly comprising terpenoids and esters. Samples were analyzed by HS-SPME using a triphase DVB/Carboxen/PDMS 50/30 μm SPME fiber (Supelco Co., Bellefonte, PA, USA) followed by gas chromatography (Thermo Fisher Scientific Trace 1300 series, USA) coupled to a mass spectrometer (Thermo Fisher Scientific ISQ series MS) equipped with a TriPlus RSH autosampler. 5 ml of degassed beer sample was placed in 20 ml vials containing 1.75 g NaCl (VWR, 27810.295). 5 µl internal standard mix was added, containing 2-heptanol (1 g/L) (Sigma-Aldrich, H3003), 4-fluorobenzaldehyde (1 g/L) (Sigma-Aldrich, 128376), 2,3-hexanedione (1 g/L) (Sigma-Aldrich, 144169) and guaiacol (1 g/L) (Sigma-Aldrich, W253200) in ethanol (Fisher Chemical, E/0650DF/C17). Each sample was incubated at 60 °C in the autosampler oven with constant agitation. After 5 min equilibration, the SPME fiber was exposed to the sample headspace for 30 min. The compounds trapped on the fiber were thermally desorbed in the injection port of the chromatograph by heating the fiber for 15 min at 270 °C.
The GC-MS was equipped with a low polarity RXi-5Sil MS column (length, 20 m; internal diameter, 0.18 mm; layer thickness, 0.18 µm; Restek, Bellefonte, PA, USA). Injection was performed in splitless mode at 320 °C, a split flow of 9 ml/min, a purge flow of 5 ml/min and an open valve time of 3 min. To obtain a pulsed injection, a programmed gas flow was used whereby the helium gas flow was set at 2.7 mL/min for 0.1 min, followed by a decrease in flow of 20 ml/min to the normal 0.9 mL/min. The temperature was first held at 30 °C for 3 min and then allowed to rise to 80 °C at a rate of 7 °C/min, followed by a second ramp of 2 °C/min till 125 °C and a final ramp of 8 °C/min with a final temperature of 270 °C.
Mass acquisition range was 33 to 550 amu at a scan rate of 5 scans/s. Electron impact ionization energy was 70 eV. The interface and ion source were kept at 275 °C and 250 °C, respectively. A mix of linear n-alkanes (from C7 to C40, Supelco Co.) was injected into the GC-MS under identical conditions to serve as external retention index markers. Identification and quantification of the compounds were performed using an in-house developed R script as described in Goelen et al. and Reher et al. 87 , 88 (for package information, see Supplementary Table S8 ). Briefly, chromatograms were analyzed using AMDIS (v2.71) 89 to separate overlapping peaks and obtain pure compound spectra. The NIST MS Search software (v2.0 g) in combination with the NIST2017, FFNSC3 and Adams4 libraries were used to manually identify the empirical spectra, taking into account the expected retention time. After background subtraction and correcting for retention time shifts between samples run on different days based on alkane ladders, compound elution profiles were extracted and integrated using a file with 284 target compounds of interest, which were either recovered in our identified AMDIS list of spectra or were known to occur in beer. Compound elution profiles were estimated for every peak in every chromatogram over a time-restricted window using weighted non-negative least square analysis after which peak areas were integrated 87 , 88 . Batch effect correction was performed by normalizing against the most stable internal standard compound, 4-fluorobenzaldehyde. Out of all 284 target compounds that were analyzed, 167 were visually judged to have reliable elution profiles and were used for final analysis.
Discrete photometric and enzymatic analysis
Discrete photometric and enzymatic analysis (Thermo Scientific TM Gallery TM Plus Beermaster Discrete Analyzer) was used to measure acetic acid, ammonia, beta-glucan, iso-alpha acids, color, sugars, glycerol, iron, pH, protein, and sulfite. 2 ml of sample volume was used for the analyses. Information regarding the reagents and standard solutions used for analyses and calibrations is included in Supplementary Table S7 and Supplementary Table S9 .
NIR analyses
NIR analysis (Anton Paar Alcolyzer Beer ME System) was used to measure ethanol. Measurements comprised 50 ml of sample, and a 10% EtOH solution was used for calibration.
Correlation calculations
Pairwise Spearman Rank correlations were calculated between all chemical properties.
Sensory dataset
Trained panel.
Our trained tasting panel consisted of volunteers who gave prior verbal informed consent. All compounds used for the validation experiment were of food-grade quality. The tasting sessions were approved by the Social and Societal Ethics Committee of the KU Leuven (G-2022-5677-R2(MAR)). All online reviewers agreed to the Terms and Conditions of the RateBeer website.
Sensory analysis was performed according to the American Society of Brewing Chemists (ASBC) Sensory Analysis Methods 90 . 30 volunteers were screened through a series of triangle tests. The sixteen most sensitive and consistent tasters were retained as taste panel members. The resulting panel was diverse in age [22–42, mean: 29], sex [56% male] and nationality [7 different countries]. The panel developed a consensus vocabulary to describe beer aroma, taste and mouthfeel. Panelists were trained to identify and score 50 different attributes, using a 7-point scale to rate attributes’ intensity. The scoring sheet is included as Supplementary Data 3 . Sensory assessments took place between 10–12 a.m. The beers were served in black-colored glasses. Per session, between 5 and 12 beers of the same style were tasted at 12 °C to 16 °C. Two reference beers were added to each set and indicated as ‘Reference 1 & 2’, allowing panel members to calibrate their ratings. Not all panelists were present at every tasting. Scores were scaled by standard deviation and mean-centered per taster. Values are represented as z-scores and clustered by Euclidean distance. Pairwise Spearman correlations were calculated between taste and aroma sensory attributes. Panel consistency was evaluated by repeating samples on different sessions and performing ANOVA to identify differences, using the ‘stats’ package (v4.2.2) in R (for package information, see Supplementary Table S8 ).
Online reviews from a public database
The ‘scrapy’ package in Python (v3.6) (for package information, see Supplementary Table S8 ). was used to collect 232,288 online reviews (mean=922, min=6, max=5343) from RateBeer, an online beer review database. Each review entry comprised 5 numerical scores (appearance, aroma, taste, palate and overall quality) and an optional review text. The total number of reviews per reviewer was collected separately. Numerical scores were scaled and centered per rater, and mean scores were calculated per beer.
For the review texts, the language was estimated using the packages ‘langdetect’ and ‘langid’ in Python. Reviews that were classified as English by both packages were kept. Reviewers with fewer than 100 entries overall were discarded. 181,025 reviews from >6000 reviewers from >40 countries remained. Text processing was done using the ‘nltk’ package in Python. Texts were corrected for slang and misspellings; proper nouns and rare words that are relevant to the beer context were specified and kept as-is (‘Chimay’,’Lambic’, etc.). A dictionary of semantically similar sensorial terms, for example ‘floral’ and ‘flower’, was created and collapsed together into one term. Words were stemmed and lemmatized to avoid identifying words such as ‘acid’ and ‘acidity’ as separate terms. Numbers and punctuation were removed.
Sentences from up to 50 randomly chosen reviews per beer were manually categorized according to the aspect of beer they describe (appearance, aroma, taste, palate, overall quality—not to be confused with the 5 numerical scores described above) or flagged as irrelevant if they contained no useful information. If a beer contained fewer than 50 reviews, all reviews were manually classified. This labeled data set was used to train a model that classified the rest of the sentences for all beers 91 . Sentences describing taste and aroma were extracted, and term frequency–inverse document frequency (TFIDF) was implemented to calculate enrichment scores for sensorial words per beer.
The sex of the tasting subject was not considered when building our sensory database. Instead, results from different panelists were averaged, both for our trained panel (56% male, 44% female) and the RateBeer reviews (70% male, 30% female for RateBeer as a whole).
Beer price collection and processing
Beer prices were collected from the following stores: Colruyt, Delhaize, Total Wine, BeerHawk, The Belgian Beer Shop, The Belgian Shop, and Beer of Belgium. Where applicable, prices were converted to Euros and normalized per liter. Spearman correlations were calculated between these prices and mean overall appreciation scores from RateBeer and the taste panel, respectively.
Pairwise Spearman Rank correlations were calculated between all sensory properties.
Machine learning models
Predictive modeling of sensory profiles from chemical data.
Regression models were constructed to predict (a) trained panel scores for beer flavors and quality from beer chemical profiles and (b) public reviews’ appreciation scores from beer chemical profiles. Z-scores were used to represent sensory attributes in both data sets. Chemical properties with log-normal distributions (Shapiro-Wilk test, p < 0.05 ) were log-transformed. Missing chemical measurements (0.1% of all data) were replaced with mean values per attribute. Observations from 250 beers were randomly separated into a training set (70%, 175 beers) and a test set (30%, 75 beers), stratified per beer style. Chemical measurements (p = 231) were normalized based on the training set average and standard deviation. In total, three linear regression-based models: linear regression with first-order interaction terms (LR), lasso regression with first-order interaction terms (Lasso) and partial least squares regression (PLSR); five decision tree models, Adaboost regressor (ABR), Extra Trees (ET), Gradient Boosting regressor (GBR), Random Forest (RF) and XGBoost regressor (XGBR); one support vector machine model (SVR) and one artificial neural network model (ANN) were trained. The models were implemented using the ‘scikit-learn’ package (v1.2.2) and ‘xgboost’ package (v1.7.3) in Python (v3.9.16). Models were trained, and hyperparameters optimized, using five-fold cross-validated grid search with the coefficient of determination (R 2 ) as the evaluation metric. The ANN (scikit-learn’s MLPRegressor) was optimized using Bayesian Tree-Structured Parzen Estimator optimization with the ‘Optuna’ Python package (v3.2.0). Individual models were trained per attribute, and a multi-output model was trained on all attributes simultaneously.
Model dissection
GBR was found to outperform other methods, resulting in models with the highest average R 2 values in both trained panel and public review data sets. Impurity-based rankings of the most important predictors for each predicted sensorial trait were obtained using the ‘scikit-learn’ package. To observe the relationships between these chemical properties and their predicted targets, partial dependence plots (PDP) were constructed for the six most important predictors of consumer appreciation 74 , 75 .
The ‘SHAP’ package in Python (v0.41.0) was implemented to provide an alternative ranking of predictor importance and to visualize the predictors’ effects as a function of their concentration 68 .
Validation of causal chemical properties
To validate the effects of the most important model features on predicted sensory attributes, beers were spiked with the chemical compounds identified by the models and descriptive sensory analyses were carried out according to the American Society of Brewing Chemists (ASBC) protocol 90 .
Compound spiking was done 30 min before tasting. Compounds were spiked into fresh beer bottles, that were immediately resealed and inverted three times. Fresh bottles of beer were opened for the same duration, resealed, and inverted thrice, to serve as controls. Pairs of spiked samples and controls were served simultaneously, chilled and in dark glasses as outlined in the Trained panel section above. Tasters were instructed to select the glass with the higher flavor intensity for each attribute (directional difference test 92 ) and to select the glass they prefer.
The final concentration after spiking was equal to the within-style average, after normalizing by ethanol concentration. This was done to ensure balanced flavor profiles in the final spiked beer. The same methods were applied to improve a non-alcoholic beer. Compounds were the following: ethyl acetate (Merck KGaA, W241415), ethyl hexanoate (Merck KGaA, W243906), isoamyl acetate (Merck KGaA, W205508), phenethyl acetate (Merck KGaA, W285706), ethanol (96%, Colruyt), glycerol (Merck KGaA, W252506), lactic acid (Merck KGaA, 261106).
Significant differences in preference or perceived intensity were determined by performing the two-sided binomial test on each attribute.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support the findings of this work are available in the Supplementary Data files and have been deposited to Zenodo under accession code 10653704 93 . The RateBeer scores data are under restricted access, they are not publicly available as they are property of RateBeer (ZX Ventures, USA). Access can be obtained from the authors upon reasonable request and with permission of RateBeer (ZX Ventures, USA). Source data are provided with this paper.
Code availability
The code for training the machine learning models, analyzing the models, and generating the figures has been deposited to Zenodo under accession code 10653704 93 .
Tieman, D. et al. A chemical genetic roadmap to improved tomato flavor. Science 355 , 391–394 (2017).
Article ADS CAS PubMed Google Scholar
Plutowska, B. & Wardencki, W. Application of gas chromatography–olfactometry (GC–O) in analysis and quality assessment of alcoholic beverages – A review. Food Chem. 107 , 449–463 (2008).
Article CAS Google Scholar
Legin, A., Rudnitskaya, A., Seleznev, B. & Vlasov, Y. Electronic tongue for quality assessment of ethanol, vodka and eau-de-vie. Anal. Chim. Acta 534 , 129–135 (2005).
Loutfi, A., Coradeschi, S., Mani, G. K., Shankar, P. & Rayappan, J. B. B. Electronic noses for food quality: A review. J. Food Eng. 144 , 103–111 (2015).
Ahn, Y.-Y., Ahnert, S. E., Bagrow, J. P. & Barabási, A.-L. Flavor network and the principles of food pairing. Sci. Rep. 1 , 196 (2011).
Article CAS PubMed PubMed Central Google Scholar
Bartoshuk, L. M. & Klee, H. J. Better fruits and vegetables through sensory analysis. Curr. Biol. 23 , R374–R378 (2013).
Article CAS PubMed Google Scholar
Piggott, J. R. Design questions in sensory and consumer science. Food Qual. Prefer. 3293 , 217–220 (1995).
Article Google Scholar
Kermit, M. & Lengard, V. Assessing the performance of a sensory panel-panellist monitoring and tracking. J. Chemom. 19 , 154–161 (2005).
Cook, D. J., Hollowood, T. A., Linforth, R. S. T. & Taylor, A. J. Correlating instrumental measurements of texture and flavour release with human perception. Int. J. Food Sci. Technol. 40 , 631–641 (2005).
Chinchanachokchai, S., Thontirawong, P. & Chinchanachokchai, P. A tale of two recommender systems: The moderating role of consumer expertise on artificial intelligence based product recommendations. J. Retail. Consum. Serv. 61 , 1–12 (2021).
Ross, C. F. Sensory science at the human-machine interface. Trends Food Sci. Technol. 20 , 63–72 (2009).
Chambers, E. IV & Koppel, K. Associations of volatile compounds with sensory aroma and flavor: The complex nature of flavor. Molecules 18 , 4887–4905 (2013).
Pinu, F. R. Metabolomics—The new frontier in food safety and quality research. Food Res. Int. 72 , 80–81 (2015).
Danezis, G. P., Tsagkaris, A. S., Brusic, V. & Georgiou, C. A. Food authentication: state of the art and prospects. Curr. Opin. Food Sci. 10 , 22–31 (2016).
Shepherd, G. M. Smell images and the flavour system in the human brain. Nature 444 , 316–321 (2006).
Meilgaard, M. C. Prediction of flavor differences between beers from their chemical composition. J. Agric. Food Chem. 30 , 1009–1017 (1982).
Xu, L. et al. Widespread receptor-driven modulation in peripheral olfactory coding. Science 368 , eaaz5390 (2020).
Kupferschmidt, K. Following the flavor. Science 340 , 808–809 (2013).
Billesbølle, C. B. et al. Structural basis of odorant recognition by a human odorant receptor. Nature 615 , 742–749 (2023).
Article ADS PubMed PubMed Central Google Scholar
Smith, B. Perspective: Complexities of flavour. Nature 486 , S6–S6 (2012).
Pfister, P. et al. Odorant receptor inhibition is fundamental to odor encoding. Curr. Biol. 30 , 2574–2587 (2020).
Moskowitz, H. W., Kumaraiah, V., Sharma, K. N., Jacobs, H. L. & Sharma, S. D. Cross-cultural differences in simple taste preferences. Science 190 , 1217–1218 (1975).
Eriksson, N. et al. A genetic variant near olfactory receptor genes influences cilantro preference. Flavour 1 , 22 (2012).
Ferdenzi, C. et al. Variability of affective responses to odors: Culture, gender, and olfactory knowledge. Chem. Senses 38 , 175–186 (2013).
Article PubMed Google Scholar
Lawless, H. T. & Heymann, H. Sensory evaluation of food: Principles and practices. (Springer, New York, NY). https://doi.org/10.1007/978-1-4419-6488-5 (2010).
Colantonio, V. et al. Metabolomic selection for enhanced fruit flavor. Proc. Natl. Acad. Sci. 119 , e2115865119 (2022).
Fritz, F., Preissner, R. & Banerjee, P. VirtualTaste: a web server for the prediction of organoleptic properties of chemical compounds. Nucleic Acids Res 49 , W679–W684 (2021).
Tuwani, R., Wadhwa, S. & Bagler, G. BitterSweet: Building machine learning models for predicting the bitter and sweet taste of small molecules. Sci. Rep. 9 , 1–13 (2019).
Dagan-Wiener, A. et al. Bitter or not? BitterPredict, a tool for predicting taste from chemical structure. Sci. Rep. 7 , 1–13 (2017).
Pallante, L. et al. Toward a general and interpretable umami taste predictor using a multi-objective machine learning approach. Sci. Rep. 12 , 1–11 (2022).
Malavolta, M. et al. A survey on computational taste predictors. Eur. Food Res. Technol. 248 , 2215–2235 (2022).
Lee, B. K. et al. A principal odor map unifies diverse tasks in olfactory perception. Science 381 , 999–1006 (2023).
Mayhew, E. J. et al. Transport features predict if a molecule is odorous. Proc. Natl. Acad. Sci. 119 , e2116576119 (2022).
Niu, Y. et al. Sensory evaluation of the synergism among ester odorants in light aroma-type liquor by odor threshold, aroma intensity and flash GC electronic nose. Food Res. Int. 113 , 102–114 (2018).
Yu, P., Low, M. Y. & Zhou, W. Design of experiments and regression modelling in food flavour and sensory analysis: A review. Trends Food Sci. Technol. 71 , 202–215 (2018).
Oladokun, O. et al. The impact of hop bitter acid and polyphenol profiles on the perceived bitterness of beer. Food Chem. 205 , 212–220 (2016).
Linforth, R., Cabannes, M., Hewson, L., Yang, N. & Taylor, A. Effect of fat content on flavor delivery during consumption: An in vivo model. J. Agric. Food Chem. 58 , 6905–6911 (2010).
Guo, S., Na Jom, K. & Ge, Y. Influence of roasting condition on flavor profile of sunflower seeds: A flavoromics approach. Sci. Rep. 9 , 11295 (2019).
Ren, Q. et al. The changes of microbial community and flavor compound in the fermentation process of Chinese rice wine using Fagopyrum tataricum grain as feedstock. Sci. Rep. 9 , 3365 (2019).
Hastie, T., Friedman, J. & Tibshirani, R. The Elements of Statistical Learning. (Springer, New York, NY). https://doi.org/10.1007/978-0-387-21606-5 (2001).
Dietz, C., Cook, D., Huismann, M., Wilson, C. & Ford, R. The multisensory perception of hop essential oil: a review. J. Inst. Brew. 126 , 320–342 (2020).
CAS Google Scholar
Roncoroni, Miguel & Verstrepen, Kevin Joan. Belgian Beer: Tested and Tasted. (Lannoo, 2018).
Meilgaard, M. Flavor chemistry of beer: Part II: Flavor and threshold of 239 aroma volatiles. in (1975).
Bokulich, N. A. & Bamforth, C. W. The microbiology of malting and brewing. Microbiol. Mol. Biol. Rev. MMBR 77 , 157–172 (2013).
Dzialo, M. C., Park, R., Steensels, J., Lievens, B. & Verstrepen, K. J. Physiology, ecology and industrial applications of aroma formation in yeast. FEMS Microbiol. Rev. 41 , S95–S128 (2017).
Article PubMed PubMed Central Google Scholar
Datta, A. et al. Computer-aided food engineering. Nat. Food 3 , 894–904 (2022).
American Society of Brewing Chemists. Beer Methods. (American Society of Brewing Chemists, St. Paul, MN, U.S.A.).
Olaniran, A. O., Hiralal, L., Mokoena, M. P. & Pillay, B. Flavour-active volatile compounds in beer: production, regulation and control. J. Inst. Brew. 123 , 13–23 (2017).
Verstrepen, K. J. et al. Flavor-active esters: Adding fruitiness to beer. J. Biosci. Bioeng. 96 , 110–118 (2003).
Meilgaard, M. C. Flavour chemistry of beer. part I: flavour interaction between principal volatiles. Master Brew. Assoc. Am. Tech. Q 12 , 107–117 (1975).
Briggs, D. E., Boulton, C. A., Brookes, P. A. & Stevens, R. Brewing 227–254. (Woodhead Publishing). https://doi.org/10.1533/9781855739062.227 (2004).
Bossaert, S., Crauwels, S., De Rouck, G. & Lievens, B. The power of sour - A review: Old traditions, new opportunities. BrewingScience 72 , 78–88 (2019).
Google Scholar
Verstrepen, K. J. et al. Flavor active esters: Adding fruitiness to beer. J. Biosci. Bioeng. 96 , 110–118 (2003).
Snauwaert, I. et al. Microbial diversity and metabolite composition of Belgian red-brown acidic ales. Int. J. Food Microbiol. 221 , 1–11 (2016).
Spitaels, F. et al. The microbial diversity of traditional spontaneously fermented lambic beer. PLoS ONE 9 , e95384 (2014).
Blanco, C. A., Andrés-Iglesias, C. & Montero, O. Low-alcohol Beers: Flavor Compounds, Defects, and Improvement Strategies. Crit. Rev. Food Sci. Nutr. 56 , 1379–1388 (2016).
Jackowski, M. & Trusek, A. Non-Alcohol. beer Prod. – Overv. 20 , 32–38 (2018).
Takoi, K. et al. The contribution of geraniol metabolism to the citrus flavour of beer: Synergy of geraniol and β-citronellol under coexistence with excess linalool. J. Inst. Brew. 116 , 251–260 (2010).
Kroeze, J. H. & Bartoshuk, L. M. Bitterness suppression as revealed by split-tongue taste stimulation in humans. Physiol. Behav. 35 , 779–783 (1985).
Mennella, J. A. et al. A spoonful of sugar helps the medicine go down”: Bitter masking bysucrose among children and adults. Chem. Senses 40 , 17–25 (2015).
Wietstock, P., Kunz, T., Perreira, F. & Methner, F.-J. Metal chelation behavior of hop acids in buffered model systems. BrewingScience 69 , 56–63 (2016).
Sancho, D., Blanco, C. A., Caballero, I. & Pascual, A. Free iron in pale, dark and alcohol-free commercial lager beers. J. Sci. Food Agric. 91 , 1142–1147 (2011).
Rodrigues, H. & Parr, W. V. Contribution of cross-cultural studies to understanding wine appreciation: A review. Food Res. Int. 115 , 251–258 (2019).
Korneva, E. & Blockeel, H. Towards better evaluation of multi-target regression models. in ECML PKDD 2020 Workshops (eds. Koprinska, I. et al.) 353–362 (Springer International Publishing, Cham, 2020). https://doi.org/10.1007/978-3-030-65965-3_23 .
Gastón Ares. Mathematical and Statistical Methods in Food Science and Technology. (Wiley, 2013).
Grinsztajn, L., Oyallon, E. & Varoquaux, G. Why do tree-based models still outperform deep learning on tabular data? Preprint at http://arxiv.org/abs/2207.08815 (2022).
Gries, S. T. Statistics for Linguistics with R: A Practical Introduction. in Statistics for Linguistics with R (De Gruyter Mouton, 2021). https://doi.org/10.1515/9783110718256 .
Lundberg, S. M. et al. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2 , 56–67 (2020).
Ickes, C. M. & Cadwallader, K. R. Effects of ethanol on flavor perception in alcoholic beverages. Chemosens. Percept. 10 , 119–134 (2017).
Kato, M. et al. Influence of high molecular weight polypeptides on the mouthfeel of commercial beer. J. Inst. Brew. 127 , 27–40 (2021).
Wauters, R. et al. Novel Saccharomyces cerevisiae variants slow down the accumulation of staling aldehydes and improve beer shelf-life. Food Chem. 398 , 1–11 (2023).
Li, H., Jia, S. & Zhang, W. Rapid determination of low-level sulfur compounds in beer by headspace gas chromatography with a pulsed flame photometric detector. J. Am. Soc. Brew. Chem. 66 , 188–191 (2008).
Dercksen, A., Laurens, J., Torline, P., Axcell, B. C. & Rohwer, E. Quantitative analysis of volatile sulfur compounds in beer using a membrane extraction interface. J. Am. Soc. Brew. Chem. 54 , 228–233 (1996).
Molnar, C. Interpretable Machine Learning: A Guide for Making Black-Box Models Interpretable. (2020).
Zhao, Q. & Hastie, T. Causal interpretations of black-box models. J. Bus. Econ. Stat. Publ. Am. Stat. Assoc. 39 , 272–281 (2019).
Article MathSciNet Google Scholar
Hastie, T., Tibshirani, R. & Friedman, J. The Elements of Statistical Learning. (Springer, 2019).
Labrado, D. et al. Identification by NMR of key compounds present in beer distillates and residual phases after dealcoholization by vacuum distillation. J. Sci. Food Agric. 100 , 3971–3978 (2020).
Lusk, L. T., Kay, S. B., Porubcan, A. & Ryder, D. S. Key olfactory cues for beer oxidation. J. Am. Soc. Brew. Chem. 70 , 257–261 (2012).
Gonzalez Viejo, C., Torrico, D. D., Dunshea, F. R. & Fuentes, S. Development of artificial neural network models to assess beer acceptability based on sensory properties using a robotic pourer: A comparative model approach to achieve an artificial intelligence system. Beverages 5 , 33 (2019).
Gonzalez Viejo, C., Fuentes, S., Torrico, D. D., Godbole, A. & Dunshea, F. R. Chemical characterization of aromas in beer and their effect on consumers liking. Food Chem. 293 , 479–485 (2019).
Gilbert, J. L. et al. Identifying breeding priorities for blueberry flavor using biochemical, sensory, and genotype by environment analyses. PLOS ONE 10 , 1–21 (2015).
Goulet, C. et al. Role of an esterase in flavor volatile variation within the tomato clade. Proc. Natl. Acad. Sci. 109 , 19009–19014 (2012).
Article ADS CAS PubMed PubMed Central Google Scholar
Borisov, V. et al. Deep Neural Networks and Tabular Data: A Survey. IEEE Trans. Neural Netw. Learn. Syst. 1–21 https://doi.org/10.1109/TNNLS.2022.3229161 (2022).
Statista. Statista Consumer Market Outlook: Beer - Worldwide.
Seitz, H. K. & Stickel, F. Molecular mechanisms of alcoholmediated carcinogenesis. Nat. Rev. Cancer 7 , 599–612 (2007).
Voordeckers, K. et al. Ethanol exposure increases mutation rate through error-prone polymerases. Nat. Commun. 11 , 3664 (2020).
Goelen, T. et al. Bacterial phylogeny predicts volatile organic compound composition and olfactory response of an aphid parasitoid. Oikos 129 , 1415–1428 (2020).
Article ADS Google Scholar
Reher, T. et al. Evaluation of hop (Humulus lupulus) as a repellent for the management of Drosophila suzukii. Crop Prot. 124 , 104839 (2019).
Stein, S. E. An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J. Am. Soc. Mass Spectrom. 10 , 770–781 (1999).
American Society of Brewing Chemists. Sensory Analysis Methods. (American Society of Brewing Chemists, St. Paul, MN, U.S.A., 1992).
McAuley, J., Leskovec, J. & Jurafsky, D. Learning Attitudes and Attributes from Multi-Aspect Reviews. Preprint at https://doi.org/10.48550/arXiv.1210.3926 (2012).
Meilgaard, M. C., Carr, B. T. & Carr, B. T. Sensory Evaluation Techniques. (CRC Press, Boca Raton). https://doi.org/10.1201/b16452 (2014).
Schreurs, M. et al. Data from: Predicting and improving complex beer flavor through machine learning. Zenodo https://doi.org/10.5281/zenodo.10653704 (2024).
Download references
Acknowledgements
We thank all lab members for their discussions and thank all tasting panel members for their contributions. Special thanks go out to Dr. Karin Voordeckers for her tremendous help in proofreading and improving the manuscript. M.S. was supported by a Baillet-Latour fellowship, L.C. acknowledges financial support from KU Leuven (C16/17/006), F.A.T. was supported by a PhD fellowship from FWO (1S08821N). Research in the lab of K.J.V. is supported by KU Leuven, FWO, VIB, VLAIO and the Brewing Science Serves Health Fund. Research in the lab of T.W. is supported by FWO (G.0A51.15) and KU Leuven (C16/17/006).
Author information
These authors contributed equally: Michiel Schreurs, Supinya Piampongsant, Miguel Roncoroni.
Authors and Affiliations
VIB—KU Leuven Center for Microbiology, Gaston Geenslaan 1, B-3001, Leuven, Belgium
Michiel Schreurs, Supinya Piampongsant, Miguel Roncoroni, Lloyd Cool, Beatriz Herrera-Malaver, Florian A. Theßeling & Kevin J. Verstrepen
CMPG Laboratory of Genetics and Genomics, KU Leuven, Gaston Geenslaan 1, B-3001, Leuven, Belgium
Leuven Institute for Beer Research (LIBR), Gaston Geenslaan 1, B-3001, Leuven, Belgium
Laboratory of Socioecology and Social Evolution, KU Leuven, Naamsestraat 59, B-3000, Leuven, Belgium
Lloyd Cool, Christophe Vanderaa & Tom Wenseleers
VIB Bioinformatics Core, VIB, Rijvisschestraat 120, B-9052, Ghent, Belgium
Łukasz Kreft & Alexander Botzki
AB InBev SA/NV, Brouwerijplein 1, B-3000, Leuven, Belgium
Philippe Malcorps & Luk Daenen
You can also search for this author in PubMed Google Scholar
Contributions
S.P., M.S. and K.J.V. conceived the experiments. S.P., M.S. and K.J.V. designed the experiments. S.P., M.S., M.R., B.H. and F.A.T. performed the experiments. S.P., M.S., L.C., C.V., L.K., A.B., P.M., L.D., T.W. and K.J.V. contributed analysis ideas. S.P., M.S., L.C., C.V., T.W. and K.J.V. analyzed the data. All authors contributed to writing the manuscript.
Corresponding author
Correspondence to Kevin J. Verstrepen .
Ethics declarations
Competing interests.
K.J.V. is affiliated with bar.on. The other authors declare no competing interests.
Peer review
Peer review information.
Nature Communications thanks Florian Bauer, Andrew John Macintosh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information, peer review file, description of additional supplementary files, supplementary data 1, supplementary data 2, supplementary data 3, supplementary data 4, supplementary data 5, supplementary data 6, supplementary data 7, reporting summary, source data, source data, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Schreurs, M., Piampongsant, S., Roncoroni, M. et al. Predicting and improving complex beer flavor through machine learning. Nat Commun 15 , 2368 (2024). https://doi.org/10.1038/s41467-024-46346-0
Download citation
Received : 30 October 2023
Accepted : 21 February 2024
Published : 26 March 2024
DOI : https://doi.org/10.1038/s41467-024-46346-0
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Translational Research newsletter — top stories in biotechnology, drug discovery and pharma.
share this!
March 27, 2024
This article has been reviewed according to Science X's editorial process and policies . Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
written by researcher(s)
English dominates scientific research—here's how we can fix it, and why it matters
by Elea Giménez Toledo, The Conversation

It is often remarked that Spanish should be more widely spoken or understood in the scientific community given its number of speakers around the world, a figure the Instituto Cervantes places at almost 600 million .
However, millions of speakers do not necessarily grant a language strength in academia. This has to be cultivated on a scientific, political and cultural level, with sustained efforts from many institutions and specialists.
The scientific community should communicate in as many languages as possible
By some estimates, as much as 98% of the world's scientific research is published in English , while only around 18% of the world's population speaks it. This makes it essential to publish in other languages if we are to bring scientific research to society at large.
The value of multilingualism in science has been highlighted by numerous high profile organizations, with public declarations and statements on the matter from the European Charter for Researchers , the Helsinki Initiative on Multiligualism , the Unesco Recommendation on Open Science , the OPERAS Multiligualism White Paper , the Latin American Forum on Research Assessment , the COARA Agreement on Reforming Research Assessment , and the Declaration of the 5th Meeting of Minsters and Scientific Authorities of Ibero-American Countries . These organizations all agree on one thing: all languages have value in scientific communication.
As the last of these declarations points out, locally, regionally and nationally relevant research is constantly being published in languages other than English. This research has an economic, social and cultural impact on its surrounding environment, as when scientific knowledge is disseminated it filters through to non-academic professionals, thus creating a broader culture of knowledge sharing.
Greater diversity also enables fluid dialogue among academics who share the same language, or who speak and understand multiple languages. In Ibero-America, for example, Spanish and Portuguese can often be mutually understood by non-native speakers, allowing them to share the scientific stage. The same happens in Spain with the majority of its co-official languages .
No hierarchies, no categories
Too often, scientific research in any language other than English is automatically seen as second tier, with little consideration for the quality of the work itself.
This harmful prejudice ignores the work of those involved, especially in the humanities and social sciences. It also profoundly undermines the global academic community's ability to share knowledge with society.
By defending and preserving multilingualism, the scientific community brings research closer to those who need it. Failing to pursue this aim means that academia cannot develop or expand its audience. We have to work carefully, systematically and consistently in every language available to us.
The logistics of strengthening linguistic diversity in science
Making a language stronger in academia is a complex process. It does not happen spontaneously, and requires careful coordination and planning. Efforts have to come from public and private institutions, the media, and other cultural outlets, as well as from politicians, science diplomacy , and researchers themselves.
Many of these elements have to work in harmony, as demonstrated by the Spanish National Research Council's work in ES CIENCIA , a project which seeks to unite scientific and and political efforts.
Academic publishing and AI models: a new challenge
The global academic environment is changing as a result the digital transition and new models of open access. Research into publishers of scientific content in other languages will be essential to understanding this shift. One thing is clear though: making scientific content produced in a particular language visible and searchable online is crucial to ensuring its strength.
In the case of academic books, the transition to open access has barely begun , especially in the commercial publishing sector, which releases around 80% of scientific books in Spain. As with online publishing, a clear understanding will make it possible to design policies and models that account for the different ways of disseminating scientific research, including those that communicate locally and in other languages. Greater linguistic diversity in book publishing can also allow us to properly recognize the work done by publishers in sharing research among non-English speakers.
Making publications, datasets, and other non-linguistic research results easy to find is another vital element, which requires both scientific and technical support. The same applies to expanding the corpus of scientific literature in Spanish and other languages, especially since this feeds into generative artificial intelligence models.
If linguistically diverse scientific content is not incorporated into AI systems, they will spread information that is incomplete, biased or misleading: a recent Spanish government report on the state of Spanish and co-official languages points out that 90% of the text currently fed into AI is written in English.
Deep study of terminology is essential
Research into terminology is of the utmost importance in preventing the use of improvised, imprecise language or unintelligible jargon. It can also bring huge benefits for the quality of both human and machine translations, specialized language teaching, and the indexing and organization of large volumes of documents.
Terminology work in Spanish is being carried out today thanks to the processing of large language corpuses by AI and researchers in the TeresIA project, a joint effort coordinated by the Spanish National Research Council. However, 15 years of ups and downs were needed to to get such a project off the ground in Spanish.
The Basque Country, Catalonia and Galicia, on the other hand, have worked intensively and systematically on their respective languages. They have not only tackled terminology as a public language policy issue, but have also been committed to established terminology projects for a long time.
Multiligualism is a global issue
This need for broader diversity also applies to Ibero-America as a whole, where efforts are being coordinated to promote Spanish and Portuguese in academia, notably by the Ibero-American General Secretariat and the Mexican National Council of Humanities, Sciences and Technologies .
While this is sorely needed, we cannot promote the region's two most widely spoken languages and also ignore its diversity of indigenous and co-official languages. These are also involved in the production of knowledge, and are a vehicle for the transfer of scientific information, as demonstrated by efforts in Spain.
Each country has its own unique role to play in promoting greater linguistic diversity in scientific communication. If this can be achieved, the strength of Iberian languages—and all languages, for that matter—in academia will not be at the mercy of well intentioned but sporadic efforts. It will, instead, be the result of the scientific community's commitment to a culture of knowledge sharing.
Provided by The Conversation
Explore further
Feedback to editors

A microbial plastic factory for high-quality green plastic
2 hours ago

Can the bias in algorithms help us see our own?
4 hours ago

Humans have converted at least 250,000 acres of estuaries to cities and farms in last 35 years, study finds
5 hours ago

Mysterious bones may have belonged to gigantic ichthyosaurs

Hurricane risk perception drops after storms hit, study shows

Peter Higgs, who proposed the existence of the 'God particle,' has died at 94

Scientists help link climate change to Madagascar's megadrought
7 hours ago

Heat from El Niño can warm oceans off West Antarctica—and melt floating ice shelves from below
8 hours ago
Peregrine falcons expose lasting harms of flame retardant use
9 hours ago

The hidden role of the Milky Way in ancient Egyptian mythology
Relevant physicsforums posts, what are your favorite disco "classics", cover songs versus the original track, which ones are better, interesting anecdotes in the history of physics.
13 hours ago
Biographies, history, personal accounts
Apr 7, 2024
Purpose of the Roman bronze dodecahedrons: are you convinced?
Apr 6, 2024
Favorite Mashups - All Your Favorites in One Place
Apr 5, 2024
More from Art, Music, History, and Linguistics
Related Stories

Prestigious journals make it hard for scientists who don't speak English to get published, study finds
Mar 23, 2024

Tracing the evolution of sign languages using computer modeling
Feb 2, 2024

Trilingual study shows how non-native languages interact with each other when multilinguals talk
Feb 16, 2023

Non-English-language science could help save biodiversity
Oct 7, 2021

Fewer than 1% of schools in England have full policies on second languages, language learning and English
Jan 17, 2024

Why all languages have words for 'this' and 'that'
Oct 30, 2023
Recommended for you
First languages of North America traced back to two very different language groups from Siberia
12 hours ago

The 'Iron Pipeline': Is Interstate 95 the connection for moving guns up and down the East Coast?
11 hours ago

Americans are bad at recognizing conspiracy theories when they believe they're true, says study
Apr 8, 2024

Earth, the sun and a bike wheel: Why your high-school textbook was wrong about the shape of Earth's orbit

Giving eyeglasses to workers in developing countries boosts income
Let us know if there is a problem with our content.
Use this form if you have come across a typo, inaccuracy or would like to send an edit request for the content on this page. For general inquiries, please use our contact form . For general feedback, use the public comments section below (please adhere to guidelines ).
Please select the most appropriate category to facilitate processing of your request
Thank you for taking time to provide your feedback to the editors.
Your feedback is important to us. However, we do not guarantee individual replies due to the high volume of messages.
E-mail the story
Your email address is used only to let the recipient know who sent the email. Neither your address nor the recipient's address will be used for any other purpose. The information you enter will appear in your e-mail message and is not retained by Phys.org in any form.
Newsletter sign up
Get weekly and/or daily updates delivered to your inbox. You can unsubscribe at any time and we'll never share your details to third parties.
More information Privacy policy
Donate and enjoy an ad-free experience
We keep our content available to everyone. Consider supporting Science X's mission by getting a premium account.
E-mail newsletter
With the planet facing a 'polycrisis', biodiversity researchers uncover major knowledge gaps
Connecting the study of infectious disease spread, biodiversity loss and climate change could offer win-win-win solutions for planetary health.
A scientific review has found almost no research studying the interconnections across three major threats to planetary health, despite UN assessments suggesting one million species are at risk of extinction, a global pandemic that resulted in over six million excess deaths, and a record-breaking year of global temperatures.
"When we began to look into it, we had suspicions the number of studies would be low, but not that low," says Dr. Jonathan Davies, a researcher with University of British Columbia's Biodiversity Research Centre who led the study, published today in The Lancet Planetary Health .
"There are misperceptions in the research community that more work in this area has already been done -- but when you look for studies investigating the mechanisms linking the three crises, there isn't much there at all."
"I believe the majority of people would prefer to live in a more sustainable and biodiverse world, and empirical data show that people are healthier and have an increased feeling of well-being when closer to nature."
In a review of over 1.8 million research articles published over the last decade, Dr. Davies and his team uncovered only a minuscule number of studies -- 128 -- investigating inter-connected drivers across infectious disease spread, biodiversity loss and climate change.
Human malaria was cited as a prime example of an emerging poly-crisis being super charged by overlapping pressures -- climate change impacting mosquito distributions, development and vectors in ways that aren't straightforward to predict.
The paper analysed research studies investing either infectious disease spread, biodiversity loss or climate change. While roughly 40,000 studies considered two of the areas in conjunction, only 505 combined research on all three areas. And only 128 actually investigated the mechanistic links connecting all three threats. And in those cases, the studies are overly focused on just three areas: infectious disease in amphibians, forest health, and Lyme disease.
The research team outlines how scientists and policy makers can better study the links and feedbacks between the crises -- making it possible to identify pathways with win-win-win outcomes and also avoiding unintended consequences of only taking action in one area, and ignoring others.
"Greater effort needs to be made to search for solutions with cross-benefits," adds Dr. Alaina Pfenning-Butterworth, who conducted the study while at UBC Botany.
"For example, planting huge numbers of new trees in order to sequesters carbon can appear like a solution to climate change, but may lead to unanticipated consequences -- such as loses of native diversity and monoculture forests that are at increased risk of disease outbreaks."
The paper also argues that despite the best efforts of the research community and funding agencies, scientists from different disciplines need to work together more closely, including veterinary schools, medical schools, ecologists, conservation biologists, and computer scientists.
"I believe the majority of people would prefer to live in a more sustainable and biodiverse world, and empirical data show that people are healthier and have an increased feeling of well-being when closer to nature," says Dr. Davies.
"But there's broad scientific consensus that 'business as usual' is unsustainable, and we risk approaching a planetary tipping point beyond which reversing course will become exponentially more difficult. We have a valuable window of opportunity to decide how our future looks."
- Environmental Awareness
- Global Warming
- Environmental Issues
- Environmental Policies
- Public Health
- World Development
- Resource Shortage
- Global climate model
- Public health
- Climate change mitigation
- Global warming controversy
- Climate engineering
- Attribution of recent climate change
- Scientific opinion on climate change
- Temperature record of the past 1000 years
Story Source:
Materials provided by University of British Columbia . Note: Content may be edited for style and length.
Journal Reference :
- Alaina Pfenning-Butterworth, Lauren B Buckley, John M Drake, Johannah E Farner, Maxwell J Farrell, Alyssa-Lois M Gehman, Erin A Mordecai, Patrick R Stephens, John L Gittleman, T Jonathan Davies. Interconnecting global threats: climate change, biodiversity loss, and infectious diseases . The Lancet Planetary Health , 2024; 8 (4): e270 DOI: 10.1016/S2542-5196(24)00021-4
Cite This Page :
Explore More
- Pregnancy Accelerates Biological Aging
- Tiny Plastic Particles Are Found Everywhere
- What's Quieter Than a Fish? A School of Them
- Do Odd Bones Belong to Gigantic Ichthyosaurs?
- Big-Eyed Marine Worm: Secret Language?
- Unprecedented Behavior from Nearby Magnetar
- Soft, Flexible 'Skeletons' for 'Muscular' Robots
- Toothed Whale Echolocation and Jaw Muscles
- Friendly Pat On the Back: Free Throws
- How the Moon Turned Itself Inside Out
Trending Topics
Strange & offbeat.
April 2, 2024
Eclipse Psychology: When the Sun and Moon Align, So Do We
How a total solar eclipse creates connection, unity and caring among the people watching
By Katie Weeman

Students observing a partial solar eclipse on June 21, 2020, in Lhokseumawe, Aceh Province, Indonesia.
NurPhoto/Getty Images
This article is part of a special report on the total solar eclipse that will be visible from parts of the U.S., Mexico and Canada on April 8, 2024.
It was 11:45 A.M. on August 21, 2017. I was in a grassy field in Glendo, Wyo., where I was surrounded by strangers turned friends, more than I could count—and far more people than had ever flocked to this town, population 210 or so. Golden sunlight blanketed thousands of cars parked in haphazard rows all over the rolling hills. The shadows were quickly growing longer, the air was still, and all of our faces pointed to the sky. As the moon progressively covered the sun, the light melted away, the sky blackened, and the temperature dropped. At the moment of totality, when the moon completely covered the sun , some people around me suddenly gasped. Some cheered; some cried; others laughed in disbelief.
Exactly 53 minutes later, in a downtown park in Greenville, S.C., the person who edited this story and the many individuals around him reacted in exactly the same ways.
On supporting science journalism
If you're enjoying this article, consider supporting our award-winning journalism by subscribing . By purchasing a subscription you are helping to ensure the future of impactful stories about the discoveries and ideas shaping our world today.
When a total solar eclipse descends—as one will across Mexico, the U.S. and Canada on April 8—everyone and everything in the path of totality are engulfed by deep shadow. Unlike the New Year’s Eve countdown that lurches across the globe one blocky time zone after another, the shadow of totality is a dark spot on Earth that measures about 100 miles wide and cruises steadily along a path, covering several thousand miles in four to five hours. The human experiences along that path are not isolated events any more than individual dominoes are isolated pillars in a formation. Once that first domino is tipped, we are all linked into something bigger—and unstoppable. We all experience the momentum and the awe together.
When this phenomenon progresses from Mexico through Texas, the Great Lakes and Canada on April 8, many observers will describe the event as life-changing, well beyond expectations. “You feel a sense of wrongness in those moments before totality , when your surroundings change so rapidly,” says Kate Russo, an author, psychologist and eclipse chaser. “Our initial response is to ask ourselves, ‘Is this an opportunity or a threat?’ When the light changes and the temperature drops, that triggers primal fear. When we have that threat response, our whole body is tuned in to taking in as much information as possible.”
Russo, who has witnessed 13 total eclipses and counting, has interviewed eclipse viewers from around the world. She continues to notice the same emotions felt by all. They begin with that sense of wrongness and primal fear as totality approaches. When totality starts, we feel powerful awe and connection to the world around us. A sense of euphoria develops as we continue watching, and when it’s over, we have a strong desire to seek out the next eclipse.
“The awe we feel during a total eclipse makes us think outside our sense of self. It makes you more attuned to things outside of you,” says Sean Goldy, a postdoctoral fellow at the department of psychiatry and behavioral sciences at Johns Hopkins University.
Goldy and his team analyzed Twitter data from nearly 2.9 million people during the 2017 total solar eclipse. They found that people within the path of totality were more likely to use not only language that expressed awe but also language that conveyed being unified and affiliated with others. That meant using more “we” words (“us” instead of “me”) and more humble words (“maybe” instead of “always”).
“During an eclipse, people have a broader, more collective focus,” Goldy says. “We also found that the more people expressed awe, the more likely they were to use those ‘we’ words, indicating that people who experience this emotion feel more connected with others.”
This connectivity ties into a sociological concept known as “collective effervescence,” Russo and Goldy say. When groups of humans come together over a shared experience, the energy is greater than the sum of its parts. If you’ve ever been to a large concert or sporting event, you’ve felt the electricity generated by a hive of humans. It magnifies our emotions.
I felt exactly that unified feeling in the open field in Glendo, as if thousands of us were breathing as one. But that’s not the only way people can experience a total eclipse.
During the 2008 total eclipse in Mongolia “I was up on a peak,” Russo recounts. “I was with only my husband and a close friend. We had left the rest of our 25-person tour group at the bottom of the hill. From that vantage point, when the shadow came sweeping in, there was not one man-made thing I could see: no power lines, no buildings or structures. Nothing tethered me to time: It could have been thousands of years ago or long into the future. In that moment, it was as if time didn’t exist.”
Giving us the ability to unhitch ourselves from time—to stop dwelling on time is a unique superpower of a total eclipse. In Russo’s work as a clinical psychologist, she notices patterns in our modern-day mentality. “People with anxiety tend to spend a lot of time in the future. And people with depression spend a lot of time in the past,” she says. An eclipse, time and time again, has the ability to snap us back into the present, at least for a few minutes. “And when you’re less anxious and worried, it opens you up to be more attuned to other people, feel more connected, care for others and be more compassionate,” Goldy says.
Russo, who founded Being in the Shadow , an organization that provides information about total solar eclipses and organizes eclipse events around the world, has experienced this firsthand. Venue managers regularly tell her that eclipse crowds are among the most polite and humble: they follow the rules; they pick up their garbage—they care.
Eclipses remind us that we are part of something bigger, that we are connected with something vast. In the hours before and after totality you have to wear protective glasses to look at the sun, to prevent damage to your eyes. But during the brief time when the moon blocks the last of the sun’s rays, you can finally lower your glasses and look directly at the eclipse. It’s like making eye contact with the universe.
“In my practice, usually if someone says, ‘I feel insignificant,’ that’s a negative thing. But the meaning shifts during an eclipse,” Russo says. To feel insignificant in the moon’s shadow instead means that your sense of self shrinks, that your ego shrinks, she says.
The scale of our “big picture” often changes after witnessing the awe of totality, too. “When you zoom out—really zoom out—it blows away our differences,” Goldy says. When you sit in the shadow of a celestial rock blocking the light of a star 400 times its size that burns at 10,000 degrees Fahrenheit on its surface, suddenly that argument with your partner, that bill sitting on your counter or even the differences among people’s beliefs, origins or politics feel insignificant. When we shift our perspective, connection becomes boundless.
You don’t need to wait for the next eclipse to feel this way. As we travel through life, we lose our relationship with everyday awe. Remember what that feels like? It’s the way a dog looks at a treat or the way my toddler points to the “blue sky!” outside his car window in the middle of rush hour traffic. To find awe, we have to surrender our full attention to the beauty around us. During an eclipse, that comes easily. In everyday life, we may need to be more intentional.
“Totality kick-starts our ability to experience wonder,” Russo says. And with that kick start, maybe we can all use our wonderment faculties more—whether that means pausing for a moment during a morning walk, a hug or a random sunset on a Tuesday. In the continental U.S., we won’t experience another total eclipse until 2044. Let’s not wait until then to seek awe and connection.
A Solar Eclipse Means Big Science
By Katrina Miller April 1, 2024
- Share full article

On April 8, cameras all over North America will make a “megamovie” of the sun’s corona, like this one from the 2017 eclipse. The time lapse will help scientists track the behavior of jets and plumes on the sun’s surface.
There’s more science happening along the path of totality →
An app named SunSketcher will help the public take pictures of the eclipse with their phones.
Scientists will use these images to study deviations in the shape of the solar surface , which will help them understand the sun’s churning behavior below.
The sun right now is approaching peak activity. More than 40 telescope stations along the eclipse’s path will record totality.
By comparing these videos to what was captured in 2017 — when the sun was at a lull — researchers can learn how the sun’s magnetism drives the solar wind, or particles that stream through the solar system.
Students will launch giant balloons equipped with cameras and sensors along the eclipse’s path.
Their measurements may improve weather forecasting , and also produce a bird’s eye view of the moon’s shadow moving across the Earth.
Ham radio operators will send signals to each other across the path of totality to study how the density of electrons in Earth’s upper atmosphere changes .
This can help quantify how space weather produced by the sun disrupts radar communication systems.
(Animation by Dr. Joseph Huba, Syntek Technologies; HamSCI Project, Dr. Nathaniel Frissell, the University of Scranton, NSF and NASA.)
NASA is also studying Earth’s atmosphere, but far from the path of totality.
In Virginia, the agency will launch rockets during the eclipse to measure how local drops in sunlight cause ripple effects hundreds of miles away . The data will clarify how eclipses and other solar events affect satellite communications, including GPS.
Biologists in San Antonio plan to stash recording devices in beehives to study how bees orient themselves using sunlight , and how the insects respond to the sudden atmospheric changes during a total eclipse.
Two researchers in southern Illinois will analyze social media posts to understand tourism patterns in remote towns , including when visitors arrive, where they come from and what they do during their visits.
Results can help bolster infrastructure to support large events in rural areas.
Read more about the eclipse:

Advertisement

IMAGES
VIDEO
COMMENTS
Step-by-step instructions for reading a primary research article. 1. Begin by reading the introduction, not the abstract. The abstract is that dense first paragraph at the very beginning of a paper. In fact, that's often the only part of a paper that many non-scientists read when they're trying to build a scientific argument.
The results and methods sections allow you to pull apart a paper to ensure it stands up to scientific rigor. Always think about the type of experiments performed, and whether these are the most appropriate to address the question proposed. Ensure that the authors have included relevant and sufficient numbers of controls.
You are new to reading scientific papers. 1: For each panel of each figure, focus particularly on the questions outlined in Rule 3. 2: ... Scientists write original research papers primarily to present new data that may change or reinforce the collective knowledge of a field. Therefore, the most important parts of this type of scientific paper ...
Content: Scientific paper format. How to read a scientific paper in 3 steps. Step 1: Identify your motivations for reading a scientific paper. Step 2: Use selective reading to gain a high-level understanding of the scientific paper. Step 3: Read straight through to achieve a deep understanding of a scientific paper.
Because scientific articles are different from other texts, like novels or newspaper stories, they should be read differently. Research papers follow the well-known IMRD format — an abstract followed by the Introduction, Methods, Results and Discussion. They have multiple cross references and tables as well as supplementary material, such as ...
One should read the title and Abstract first to establish a blueprint for what the author(s) wants to convey related to their research. The next step in reading a manuscript will depend upon one's prior knowledge of the topic, goals of reading the paper, level of concentration/time to devote to reading, and overall interest.
Step 1: find. I used to find new papers by aimlessly scrolling through science Twitter. But because I often got distracted by irrelevant tweets, that wasn't very efficient. I also signed up for ...
It may help you to familiarize yourself with the 10 Stages of Reading a Scientific Paper: 1. Optimism. "This can't be too difficult," you tell yourself with a smile—in the same way you tell yourself, "It's not damaging to drink eight cups of coffee a day" or "There are plenty of tenure-track jobs." After all, you've been reading words for ...
1. The title. A one-liner, should convey the main message of the paper. 2. The abstract summarizes the main points of the paper. It should have a few sentences to introduce the problem, followed by the main results and a conclusion. The abstract is meant to generate interest in the paper, also from scientists who are not directly familiar with ...
Researchers must read papers for several reasons: to re-view them for a conference or a class, to keep current in their eld, or for a literature survey of a new eld. A typi-cal researcher will likely spend hundreds of hours every year reading papers. Learning to e ciently read a paper is a critical but rarely taught skill.
Make sure to read the accompanying figure legend so you know what all the variables are, and refer back to the methods if you're unsure of how the data was collected. Try to analyze and draw your own conclusions from the figures. Then, once you've looked at all the figures, go back and read the results text.
Attempting to read a scientific or scholarly research article for the first time may seem overwhelming and confusing. This guide details how to read a scientific article step-by-step. First, you should not approach a scientific article like a textbook— reading from beginning to end of the chapter or book without pause for reflection or criticism.
Start with the broad and then to the specific. Begin by understanding the topic of the article before trying to dig through all the fine points the author is making. Always read the tables, charts, and figures. These will give a visual clue to the methods and results sections of the paper and help you to understand the data.
You might have tried to read scientific papers before and been frustrated by the dense, stilted writing and the unfamiliar jargon. I remember feeling this way! Reading and understanding research papers is a skill which every single doctor and scientist has had to learn during graduate school. You can learn it too, but like any skill it takes ...
Step 4: Focus on the Figures. If you want to read a scientific paper effectively, the results section is where you should spend most of your time. This is because the results are the meat of the paper, without which the paper has no purpose. How you "read" the results is important because while the text is good to read, it is just a ...
What is a scientific paper? You might be wondering how a scientific paper differs from an article you might read in a newspaper or a book. Scientific papers are usually written by teams of scientists (rarely a single scientist) are are meant to either (1) present new data (known as a research paper) or (2) summarize existing literature (known as a review paper).
Skim All the Sections of the Paper. Read the Introduction. Identify How the Paper Fits into the Field of Research. Read the Discussion Section. Read the Abstract. Read the Methods and Results Section. 1. Always Read the Disclosure Section. This section is crucial to decipher whether the study is biased.
Building on past knowledge, the reader should select papers about which he already holds an opinion. Rather than starting at the beginning, this author suggests approaching a paper by reading the conclusions in the abstract first. The methods should be next reviewed, then the results--first in the abstract, and then the full paper.
Organization of Research Papers Research papers are rigidly constructed. Science editors require that submitted papers conform to universal guide-lines and style. This is fortunate for the reader, as this predictable organization allows a consistent approach to reading and evaluating a research paper. All research pa-
Search Millions of Research Papers. This fulltext search index includes over 35 million research articles and other scholarly documents preserved in the Internet Archive. The collection spans from digitized copies of eighteenth century journals through the latest Open Access conference proceedings and preprints crawled from the World Wide Web.
Notably, 46.2% of established/leading researchers or research managers read the Results-text section fourth (in IMRAD order), compared to 29.4% of MSc by research/PhD students and 28.2% of mid-career researchers who did so. Similarly, a higher percentage of established/leading researchers or research managers indicated reading the Results ...
How to read a scientific research paper. C. Durbin. Published in Respiratory care 1 October 2009. Medicine. TLDR. Rather than starting at the beginning, the author suggests approaching a paper by reading the conclusions in the abstract first, then reviewing the methods and the results first, and then the full paper. Expand.
Having good habits for reading scientific literature is key to setting oneself up for success, identifying new research questions, and filling in the gaps in one's current understanding; developing these good habits is the first crucial step. Advice typically centers around two main tips: read actively and read often.
The perception and appreciation of food flavor depends on many interacting chemical compounds and external factors, and therefore proves challenging to understand and predict. Here, we combine ...
It is often remarked that Spanish should be more widely spoken or understood in the scientific community given its number of speakers around the world, a figure the Instituto Cervantes places at ...
The paper analysed research studies investing either infectious disease spread, biodiversity loss or climate change. While roughly 40,000 studies considered two of the areas in conjunction, only ...
Lixisenatide, a glucagon-like peptide-1 receptor agonist used for the treatment of diabetes, has shown neuroprotective properties in a mouse model of Parkinson's disease.In this phase 2, double ...
The U.S. National Science Foundation (NSF) is spending $571 million to build the Vera C. Rubin Observatory in Chile so astronomers can survey the sky in unprecedented detail for evidence of dark matter and energy. It's part of the agency's mission to fund basic research.
This article is part of a special report on the total solar eclipse that will be visible from parts of the U.S., Mexico and Canada on April 8, 2024. It was 11:45 A.M. on August 21, 2017. I was in ...
On April 8, cameras all over North America will make a "megamovie" of the sun's corona, like this one from the 2017 eclipse. The time lapse will help scientists track the behavior of jets ...