Volume 21 Supplement 9

Selected Articles from the 20th International Conference on Bioinformatics & Computational Biology (BIOCOMP 2019)
- Introduction
- Open access
- Published: 03 December 2020
Current trend and development in bioinformatics research
- Yuanyuan Fu 1 ,
- Zhougui Ling 1 , 2 ,
- Hamid Arabnia 3 &
- Youping Deng 1
BMC Bioinformatics volume 21 , Article number: 538 ( 2020 ) Cite this article
10k Accesses
16 Citations
4 Altmetric
Metrics details
This is an editorial report of the supplements to BMC Bioinformatics that includes 6 papers selected from the BIOCOMP’19—The 2019 International Conference on Bioinformatics and Computational Biology. These articles reflect current trend and development in bioinformatics research.
The supplement to BMC Bioinformatics was proposed to launch during the BIOCOMP’19—The 2019 International Conference on Bioinformatics and Computational Biology held from July 29 to August 01, 2019 in Las Vegas, Nevada. In this congress, a variety of research areas was discussed, including bioinformatics which was one of the major focuses due to the rapid development and requirement of using bioinformatics approaches in biological data analysis, especially for omics large datasets. Here, six manuscripts were selected after strict peer review, providing an overview of the bioinformatics research trend and its application for interdisciplinary collaboration.
Cancer is one of the leading causes of morbidity and mortality worldwide. There exists an urgent need to identify new biomarkers or signatures for early detection and prognosis. Mona et al. identified biomarker genes from functional network based on the 407 differential expressed genes between lung cancer and healthy populations from a public Gene Expression Omnibus dataset. The lower expression of sixteen gene signature is associated with favorable lung cancer survival, DNA repair, and cell regulation [ 1 ]. A new class of biomarkers such as alternative splicing variants (ASV) have been studied in recent years. Various platforms and methods, for example, Affymetrix Exon-Exon Junction Array, RNA-seq, and liquid chromatography tandem mass spectrometry (LC–MS/MS), have been developed to explore the role of ASV in human disease. Zhang et al. have developed a bioinformatics workflow to combine LC–MS/MS with RNA-seq which provide new opportunities in biomarker discovery. In their study, they identified twenty-six alternative splicing biomarker peptides with one single intron event and one exon skipping event; further pathways indicated the 26 peptides may be involved in cancer, signaling, metabolism, regulation, immune system and hemostasis pathways which validated by the RNA-seq analysis [ 2 ].
Proteins serve crucial functions in essentially all biological processes and the function directly depends on their three-dimensional structures. Traditional approaches to elucidation of protein structures by NMR spectroscopy are time consuming and expensive, however, the faster and more cost-effective methods are critical in the development of personalized medicine. Cole et al. improved the REDRAFT software package in the important areas of usability, accessibility, and the core methodology which resulted in the ability to fold proteins [ 3 ].
The human microbiome is the aggregation of microorganisms that reside on or within human bodies. Rebecca et al. discussed the tissue-associated microbial detection in cancer using next generation sequencing (NGS). Various computational frameworks could shed light on the role of microbiota in cancer pathogenesis [ 4 ]. How to analyze the human microbiome data efficiently is a huge challenge. Zhang et al. developed a nonparametric test based on inter-point distance to evaluate statistical significance from a Bayesian point of view. The proposed test is more efficient and sensitive to the compositional difference compared with the traditional mean-based method [ 5 ].
Human disease is also considered as the cause of the interaction between genetic and environmental factors. In the last decades, there was a growing interest in the effect of metal toxicity on human health. Evaluating the toxicity of chemical mixture and their possible mechanism of action is still a challenge for humans and other organisms, as traditional methods are very time consuming, inefficient, and expensive, so a limited number of chemicals can be tested. In order to develop efficient and accurate predictive models, Yu et al. compared the results among a classification algorithm and identified 15 gene biomarkers with 100% accuracy for metal toxicant using a microarray classifier analysis [ 6 ].
Currently, there is a growing need to convert biological data into knowledge through a bioinformatics approach. We hope these articles can provide up-to-date information of research development and trend in bioinformatics field.
Availability of data and materials
Not applicable.
Abbreviations
The 2019 International Conference on Bioinformatics and Computational Biology
Liquid chromatography tandem mass spectrometry
Alternative splicing variants
Nuclear Magnetic Resonance
Residual Dipolar Coupling based Residue Assembly and Filter Tool
Next generation sequencing
Mona Maharjan RBT, Chowdhury K, Duan W, Mondal AM. Computational identification of biomarker genes for lung cancer considering treatment and non-treatment studies. 2020. https://doi.org/10.1186/s12859-020-3524-8 .
Zhang F, Deng CK, Wang M, Deng B, Barber R, Huang G. Identification of novel alternative splicing biomarkers for breast cancer with LC/MS/MS and RNA-Seq. Mol Cell Proteomics. 2020;16:1850–63. https://doi.org/10.1186/s12859-020-03824-8 .
Article Google Scholar
Casey Cole CP, Rachele J, Valafar H. Increased usability, algorithmic improvements and incorporation of data mining for structure calculation of proteins with REDCRAFT software package. 2020. https://doi.org/10.1186/s12859-020-3522-x .
Rebecca M, Rodriguez VSK, Menor M, Hernandez BY, Deng Y. Tissue-associated microbial detection in cancer using human sequencing data. 2020. https://doi.org/10.1186/s12859-020-03831-9 .
Qingyang Zhang TD. A distance based multisample test for high-dimensional compositional data with applications to the human microbiome . 2020. https://doi.org/10.1186/s12859-020-3530-x .
Yu Z, Fu Y, Ai J, Zhang J, Huang G, Deng Y. Development of predicitve models to distinguish metals from non-metal toxicants, and individual metal from one another. 2020. https://doi.org/10.1186/s12859-020-3525-7 .
Download references
Acknowledgements
This supplement will not be possible without the support of the International Society of Intelligent Biological Medicine (ISIBM).
About this supplement
This article has been published as part of BMC Bioinformatics Volume 21 Supplement 9, 2020: Selected Articles from the 20th International Conference on Bioinformatics & Computational Biology (BIOCOMP 2019). The full contents of the supplement are available online at https://bmcbioinformatics.biomedcentral.com/articles/supplements/volume-21-supplement-9 .
Publication of this supplement has been supported by NIH grants R01CA223490 and R01 CA230514 to Youping Deng and 5P30GM114737, P20GM103466, 5U54MD007601 and 5P30CA071789.
Author information
Authors and affiliations.
Department of Quantitative Health Sciences, John A. Burns School of Medicine, University of Hawaii at Manoa, Honolulu, HI, 96813, USA
Yuanyuan Fu, Zhougui Ling & Youping Deng
Department of Pulmonary and Critical Care Medicine, The Fourth Affiliated Hospital of Guangxi Medical University, Liuzhou, 545005, China
Zhougui Ling
Department of Computer Science, University of Georgia, Athens, GA, 30602, USA
Hamid Arabnia
You can also search for this author in PubMed Google Scholar
Contributions
YF drafted the manuscript, ZL, HA, and YD revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Correspondence to Youping Deng .
Ethics declarations
Ethics approval and consent to participate, consent for publication, competing interests.
The authors declare that they have no competing interests.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Fu, Y., Ling, Z., Arabnia, H. et al. Current trend and development in bioinformatics research. BMC Bioinformatics 21 (Suppl 9), 538 (2020). https://doi.org/10.1186/s12859-020-03874-y
Download citation
Published : 03 December 2020
DOI : https://doi.org/10.1186/s12859-020-03874-y
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Bioinformatics
- Human disease
BMC Bioinformatics
ISSN: 1471-2105
- General enquiries: [email protected]
Loading metrics
Open Access
Bioinformatics Projects Supporting Life-Sciences Learning in High Schools
Affiliation Instituto Gulbenkian de Ciência, Oeiras, Portugal
Affiliation Escola Secundária Stuart de Carvalhais, Queluz, Portugal
* E-mail: [email protected]
- Isabel Marques,
- Paulo Almeida,
- Renato Alves,
- Maria João Dias,
- Ana Godinho,
- José B. Pereira-Leal

Published: January 23, 2014
- https://doi.org/10.1371/journal.pcbi.1003404
- Reader Comments
The interdisciplinary nature of bioinformatics makes it an ideal framework to develop activities enabling enquiry-based learning. We describe here the development and implementation of a pilot project to use bioinformatics-based research activities in high schools, called “Bioinformatics@school.” It includes web-based research projects that students can pursue alone or under teacher supervision and a teacher training program. The project is organized so as to enable discussion of key results between students and teachers. After successful trials in two high schools, as measured by questionnaires, interviews, and assessment of knowledge acquisition, the project is expanding by the action of the teachers involved, who are helping us develop more content and are recruiting more teachers and schools.
Citation: Marques I, Almeida P, Alves R, Dias MJ, Godinho A, Pereira-Leal JB (2014) Bioinformatics Projects Supporting Life-Sciences Learning in High Schools. PLoS Comput Biol 10(1): e1003404. https://doi.org/10.1371/journal.pcbi.1003404
Editor: Fran Lewitter, Whitehead Institute, United States of America
Copyright: © 2014 Marques et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Funding: This work was funded by the Instituto Gulbenkian de Ciência. The funders had no role in the preparation of the manuscript.
Competing interests: The authors have declared that no competing interests exist.
Background and Motivation
Our lives are increasingly touched by science and technology, from the everyday activities of browsing the internet, taking a prescription drug, etc., to major societal discussions involving, for example, genetically modified foods, cloning, or stem cells. It is therefore imperative that we engage young people in science. We witnessed in the past shrinking numbers of students choosing science degrees for their university education [1] . This trend seems, however, to have been inverted both in Europe and in the United States [2] , [3] . A recent study points to the development of new and more attractive curricula and teaching methods as the driver for this increased interest [3] . In light of the growing evidence of a direct link between attitudes towards science and the way science is taught [1] , there is increasing recognition of the need to couple the traditional teacher-centred “deductive approach” to the learner-centred “inductive approach,” relying on observation, experimentation, and teacher guidance in constructing students' knowledge. This “bottom-up” approach, called enquiry-based learning (also known as problem-based learning or case-based learning) [4] recapitulates the scientific process (raising questions, collecting data, reasoning, reviewing evidence, drawing conclusions, and discussing results), thus promoting both ideas of science (scientific concepts) and ideas about science (process, practices, and critical thinking), i.e., about the Nature of Science (NOS).
Bioinformatics is a discipline at the intersection of biology, computer science, information science, mathematics, and to some extent also of chemistry and physics. It developed in response to the increasingly complex data types and relationships in biological research, addressing the need to manage and interpret biological information. This interdisciplinary nature makes bioinformatics an ideal framework to engage high school students, as it illustrates the interplay between different scientific areas, while touching on many aspects that are relevant to the younger generations—health, environment, etc. This has been recognized by many others who have implemented bioinformatics-training programs. Examples are a web-based, problem-oriented approach aimed at introducing students to bioinformatics [5] and the use of bioinformatics activities as a way to teach evolution [6] or notions of polymorphisms in the context of human genetic variation and disease [7] . Bioinformatics has also integrated with wet-lab activities in initiatives like the student-aimed “Cus-Mi-Bio” project [8] , which include gene finding activities, or in projects aimed at high school and college teachers, such as the ones at the Dolan DNA learning centre of Cold Spring Harbor Laboratory involving plant genome annotation [9] . More recently, activities that aim to introduce high school students to bioinformatics itself have also been reported [10] , and, as of 2012, an exercise using Basic Local Alignment Search Tool (BLAST) has been included on the Advanced Placement, high school biology, national test in the US ( http://apcentral.collegeboard.com/apc/members/courses/teachers_corner/218954.html ). Note, however, that these are likely isolated cases rather than the norm, as a survey revealed that in 2008 bioinformatics was still absent from the classroom in the US [11] , and likely elsewhere.
The “Bioinformatics@school” Program
We run a Bioinformatics Core at the Instituto Gulbenkian de Ciência, in Portugal, that has long been engaged in outreach activities. In 2007, we decided to implement a genomics/bioinformatics activity that would enable enquiry-based learning; link to the national curricula in biology in secondary education; introduce students to bioinformatics, genomics, and molecular biology, areas that underlie many of the key debates and products in our societies; foster active learning, making use of technologies that younger generations are increasingly comfortable with; and help teachers incorporate the latest advances in science into their teaching. We developed a prototype system that we describe the following components of here: its development, implementation, and the results of nearly five years of activity.
We developed and implemented a framework for the use of bioinformatics-based research projects in high schools to support the life-sciences curricula, which we named, in Portuguese, “Bioinformática na escola,” loosely translating to “Bioinformatics@school.” It consists of research projects that may be conducted independently by high school students of different ages, either under direct teacher supervision or as homework. Each work unit in a research project is designed to be carried out in 90 minutes, which is a standard class length in Portuguese high schools. We implemented it as a web portal ( Figure 1A, 1B )— www.bioinformatica-na-escola.org . Although primarily written in Portuguese, the site makes use of external, freely accessible bioinformatics tools and databases available in English. This is not a problem for Portuguese high school students that typically start learning English at the age of nine. Because of the dependency on external sites, we have ensured that students are given alternative access to any data on which progression to the following activity depends.
- PPT PowerPoint slide
- PNG larger image
- TIFF original image
( A ) Screenshots of the home page. ( B ) Screenshots of exercises pages.
https://doi.org/10.1371/journal.pcbi.1003404.g001
The whole program is structured as a set of projects with open-ended questions. A project may have a single activity or several, each having focused questions. Answering these focused questions enables students to discuss and/or solve the project's main question. Individual activities in the multi-activity projects were designed to also be used independently (discussed below). The concept lends itself both to classroom use, individually or in pairs, or as homework. We designed individual activities to explore specific concepts that are part of the school curriculum and the projects to be coherent with the curriculum of specific age groups, with the active collaboration of teachers in choosing the topics.
Projects are organized as follows. Once a project is selected, the student has access to a page that summarizes the problem to be solved and a link to the first activity. As the student enters one activity, s/he is presented with a sequential series of pages, each giving some background information on the specific problem the student has to follow and a brief description of the bioinformatics resources/tools to be used. At the end of each activity, the student is taken to a summary page (“now you know that…”) with an overview of the basic concepts that were addressed in the activity. All pages include links to additional information on key concepts, mostly on Wikipedia ( www.wikipedia.org ), including explanations about the resources and algorithms used in the analysis. Once the activity/final activity is complete, the student is taken to a summary page that reviews the key concepts of the project as a whole and a series of questions that act as primers for discussion amongst students and with the teacher(s) (see Figure 1A, 1B for screenshots). Table 1 summarizes the questions, concepts, and software and resources that are covered in each individual activity of “Vision,” the first multi-activity project that we have implemented in the Bioinformatics@school portal (further detailed in Text S1 ). Its implementation in schools is discussed below.
https://doi.org/10.1371/journal.pcbi.1003404.t001
Implementing “Bioinformatics@school”
Iterative development of project modules.
We started to develop “Bioinformatics@school” as a pilot project in 2007, in close collaboration with high school students and teachers. The first stage of the project consisted of identifying the topics within the high school curricula that would be amenable to bioinformatics treatment, as well as the ideal school year for the pilot to be developed. We chose 12th grade biology, in the last year of high school in the Portuguese educational system, as their curricula included multiple themes that were ideal to address using bioinformatics (such as genes, genomes, genetics, evolution, mutation, etc.), and these students would all have had several years of English language schooling (discussed above). The next phase of the project consisted in the enrollment of schools. Two secondary schools located in the Lisbon area were recruited, representing two different demographics. Escola Secundária Miguel Torga (ESMT), in Queluz, is a large suburban school that covers a variety of social strata, while Escola Secundária Quinta do Marquês (ESQM), in Oeiras, is located in a high income area with high levels of graduates and post-graduates. We engaged seven 12th grade Biology teachers, two from ESMQ and five from ESMT. One hundred and fifty students were involved in this initial pilot phase, representing multiple science-related career ambitions, ranging from engineering, health, biology, psychology, sports, etc.
We conceived the general framework described above and developed the first full project consisting of five activities, aimed at understanding how animals see different colours (“Vision” project, Table 1 ). We chose this question because we believed it to be sufficiently intriguing and relevant to engage the students (natural variants cause differential colour perception between species and between different people), but also for practical reasons—the biology of light detection via opsins is well understood, as is the 3D structure of opsins. The aim of the project is to motivate a discussion about evolution, molecular mechanisms, and disease, all inferred from bioinformatics analysis, while helping teachers and students engage with specific topics of the Life Sciences curriculum via the individual bioinformatics activities.
An innovative aspect of this project was the collaboration between scientists, teachers, and students on different aspects of the development, implementation, and testing—a three-way dialogue with continual updating in response to feedback of students and teachers. The development was iterative, first within our Bioinformatics Unit, and then in discussions with teachers. Once a first prototype was in place, one of us (IM) went to the schools to guide the students in the first activities of the project, with the help of the teacher. Student feedback was then used to improve the activities, in terms of rationale, language, and presentation.
Teacher training
Keeping up to date with the rapid developments in genomics and bioinformatics represents a challenge for high school teachers, particularly when many may have completed their training decades ago. In fact, in our experience, bioinformatics is a novel subject area to most Portuguese high school teachers. This led us to implement a parallel teacher-training program, again co-developed with the first set of teachers. Teachers were trained by bioinformatics experts, with the main goal of training to guide the students in the bioinformatics-based projects and to understand the basics of the bioinformatics methods and resources underlying each activity. We developed a teacher's manual that described the activities step by step and provided additional background information for the teacher to be comfortable with all the concepts in each activity. The teacher training consisted of having the teachers follow the same activities as the students, with the help of the teacher manual and under the supervision of a bioinformatician. We have expanded the teacher training to include seminars about applications of bioinformatics to human health, biotechnology, etc. A typical teacher training program lasts about 25 hours.
Extending the Program and Sustainability
After the successful pilot stage in 2007 the project has expanded to other geographical areas of Portugal. Thirty three new schools have joined the program, some via previously engaged teachers who took the program with them when they moved to a new school, others by new teachers who contacted us, after hearing about the project, and asked us to help them implement it in their schools. In total, schools of 11 municipalities in four Portuguese districts are currently following the program ( Figure 2A ). On their own initiative, some teachers have adapted the individual activities within the “Vision” project for use with younger students. They have also picked individual or subsets of activities and re-used them with different genes/systems, combining them in novel ways, to create new projects. They have also engaged with us to develop new projects (“Tasting Bitter”) and activities (“Tree of Life”). Furthermore, teachers are recruiting and training new teachers to use our activities. Interestingly, we observed that teachers tailored the activities to their own teaching style, some engaging the students almost at every mouse click, whereas others would only focus on explaining the basic ideas at the beginning and then discussing the outcomes at the end.
( A ) Map of schools participating, coloured by year of joining the project. ( B ) Summary of responses to confidential questionnaire. ( C ) Knowledge acquisition—each dot represents one class and the average score that students in that class achieved in the test before and after finishing the “Vision” project. ( D ) Confidence—each dot represents one class and the percent of answers that students in that class answered True or False, as opposed to answering “I don't know,” before and after finishing the “Vision” project.
https://doi.org/10.1371/journal.pcbi.1003404.g002
One aspect that worried us from early on was how to motivate teachers to engage with projects like ours when they are overwhelmed with teaching and administrative work. We realized that certification of the training is important for career progression within the Portuguese public educational system. We invested in having the project certified for teachers' continuous professional development by the national educational authorities (Conselho Científico-Pedagógico da Formação Contínua), thus making engagement with Bioinformatics@school even more appealing to the teachers. Recently we established a partnership with a teacher training centre (Centro de Formação Lezíria - Oeste) to enable other teachers in another Portuguese region to receive training in Bioinformatics activities and further promote the decentralization of “Bioinformatics@school.”
We have, thus, reason to believe that the use of the Bioinformatics@school platform is spreading on its own, with a dynamic beyond the ability of the small staff at the Bioinformatics Core that developed it.
Impact Assessment
We wished to evaluate how students and teachers perceive the program and to what extent it is an effective learning tool. These are independent questions that we addressed using different approaches. Conversations with students participating in the program suggested that they were motivated to participate in “hands-on” activities We implemented a simple confidential questionnaire to capture students' views beyond anecdotal opinions, that was given to 150 students (two schools, seven classes), during the implementation phase of the project. The results are shown in Figure 2B and reveal that the majority of the students found the approach used in this project more motivating than traditional teaching methods (58%), and enjoyed participating in it (60%). About 80% considered it had not been a waste of time and 80% would recommend the project to next year's colleagues. This type of questionnaire is useful in gauging attitudes towards the program, but it has caveats, namely that the students at this stage were very involved with the development of the Bioinformatics@school project and may be overly positive because of that. In addition, it gives no information about student learning. To address this, we devised a simple test on the concepts explored in the program, with “True/False/I Don't Know” answers ( Table S1 ). We asked four classrooms to take the test before and after the activities (this test was irrelevant for their grades). Plotting the percentage of correct answers per student before and after the activities ( Figure 2C ) revealed a dramatic increase in the proportion of correct answers, indicating that students actually gain knowledge. One surprising result was that the students appeared more confident after doing the activities: they increasingly answered the test questions as false or true, rarely using “I don't know” ( Figure 2D ). Since most of the concepts in our activities are part of the school curricula and were being covered in class by their teachers, we speculate that the decrease in “I don't know” answers may indicate that students are less afraid of venturing answers to scientific questions after doing the activities. Fear of science (“too complicated”) has been pointed out as a reason for the decreasing number of students pursuing scientific degrees [1] . This is an exciting finding that we will need to specifically evaluate further in the future. Regarding the teachers, we developed the whole program in close collaboration with them and obtained continuous feedback on the content and presentation. Although we have not as yet conducted a systematic evaluation of teachers' views about the program, the continuous contact with the currently more than seventy teachers involved suggests to us that this is a useful teaching/learning tool. In particular, teachers mention that these activities allow them to overcome the lack of laboratory-based practicals associated with some of the content in the curricula, like genetics and molecular biology. The fact that the program is spreading, with new teachers and schools recruited by word of mouth by the teachers themselves, underscores its interest and usefulness to teachers.
Discussion and Future Directions
In summary, we implemented a set of bioinformatics multi-activity research projects designed to enable enquiry-based learning in high schools. Assessment of this project has shown that students find it enjoyable and teachers believe it to be useful as a teaching aid. Objective assessment of knowledge acquisition revealed a clear positive effect both in knowledge and confidence of the students. Teachers have taken the initiative to adapt the activities to their own teaching settings and are also recruiting other teachers, which gives us further confidence in the usefulness of this project.
We have focused the projects on addressing specific biological questions, to serve the Life Sciences curriculum. This means that we don't explore the algorithmic or technological side of bioinformatics. For the future, we hope to engage teachers from mathematics, information technology, physics, and chemistry to develop projects that can serve the curricula of those particular subjects.
Recently, Form and Lewitter proposed a simple set of ten rules to guide the use of bioinformatics in high schools [12] . While these were not available at the time we were developing this project, it is interesting to note that we independently “discovered” several of these principles. We implemented individual activities with clear, simple goals (rule 1) that built on each other (rule 4), enabling students to “discover” concepts on their own (rules 5 and 8). Throughout this project we were always mindful that these activities need to serve the pre-existing curricula (Rule 3). In the future we would like to have multiple projects serving the same concepts that would allow students in each class to choose an individual project (rule 6: personalization) that they could then present and contrast to other projects pursued by their colleagues (rule 10: produce a product). We would like to develop a mapping of activities to concepts in the curricula, so that it becomes even easier for teachers to mix and match the individual activities to different contexts, thus using our project as a means to empower the teachers. Based on our experience in setting up this program, we would like to suggest two additional “simple rules” that we believe to be important when developing contents to be used in high schools:
- Engage teachers and students in the development of the activities, as a means of empowering them and ensuring that the end product meets all the cognitive and pedagogical requirements (e.g., engage the teachers in choosing the specific topics of the curricula that would benefit from bioinformatics-based projects as well as to advise on time or practical constraints on their use in the school setting; engage both teachers and students to identify weak/unappealing points in the contents and formats of the activities and to suggest better solutions, etc.).
- Evaluate the impact of the activities on engagement/enthusiasm for science and, in particular, on knowledge acquisition, as demonstrated effectiveness is the best way to get bioinformatics into the classroom. In our opinion, perpetuating useless activities just for the sake of their perceived modernity is more likely to harm the use of bioinformatics as a tool for high school science education than to advance it.
Our program was developed in Portuguese as it is targeted at Portuguese students. While this gives us potential access to a universe of more than 200 million Portuguese speakers worldwide, it is hard to use by speakers of other languages. We have started translating the whole set of activities into English, thus making Bioinformatics@school accessible to a much larger target audience. Equally, besides developing novel activities, we would like to adapt those from successful experiments elsewhere, and in due time will contact their authors directly. In this regard, the existence of a central repository of bioinformatics exercises to be used in high schools, with clear explanations according to pre-defined standards and mapping to specific concepts, would facilitate the adoption of bioinformatics in high schools. Developing standards and repositories should come naturally to the bioinformatics community!
Supporting Information
Questionnaire for impact assessment.
https://doi.org/10.1371/journal.pcbi.1003404.s001
Activities in the “Vision” project.
https://doi.org/10.1371/journal.pcbi.1003404.s002
Acknowledgments
We wish to thank all the high school teachers who have engaged with the Bioinformatics@school project, in particular Lurdes Louro (ESMT, Queluz), Filomena Delgado (ESQM, Oeiras), and Teresa Palma (Escola Secundária de Camões [ESdeC], Lisboa). We wish also to thank for their generosity and enthusiasm the initial batch of students from ESQM and ESMT who helped us develop ever better activities. Finally, we thank João Garcia and Gil Neto at the IGC, who provided invaluable IT support. We also wish to thank the Instituto Gulbenkian de Ciência for hosting this program.
- View Article
- Google Scholar
- 2. Kang K (2012) Graduate Enrollment in Science and Engineering Grew Substantially in the Past Decade but Slowed in 2010. National Center for Science and Engineering Studies. Available: http://www.nsf.gov/statistics/infbrief/nsf12317/ . Accessed 20 December 2013.
- 3. Kearney C (2010) Efforts to Increase Students' Interest in Pursuing Mathematics, Science and Technology Studies and Careers. Wastiau P, Gras-Velázquez A, Grečnerová B, Baptista R, editors. Brussels: European Schoolnet. Available: http://cms.eun.org/shared/data/pdf/spice_kearney_mst_report_nov2010.pdf . Accessed 20 December 2013.
An official website of the United States government
Here's how you know
Official websites use .gov A .gov website belongs to an official government organization in the United States.
Secure .gov websites use HTTPS. A lock ( Lock Locked padlock ) or https:// means you've safely connected to the .gov website. Share sensitive information only on official, secure websites.

Innovation: Bioinformatics
View guidelines, important information for proposers.
All proposals must be submitted in accordance with the requirements specified in this funding opportunity and in the NSF Proposal & Award Policies & Procedures Guide (PAPPG) that is in effect for the relevant due date to which the proposal is being submitted. It is the responsibility of the proposer to ensure that the proposal meets these requirements. Submitting a proposal prior to a specified deadline does not negate this requirement.
Supports research on the development of bioinformatics approaches to advance biological research in all areas funded by the Directorate for Biological Sciences.
The Bioinformatics Programmatic Area supports the design of novel and innovative bioinformatics approaches that have the potential to become part of the cyberinfrastructure that will advance or transform biological understanding and that has the potential to be broadly applicable in biology. Proposed projects may be focused on any biological process that will be better understood through the development of computational tools and approaches to acquire, model, or analyze biological information. The scope of the proposed project should include one discrete or several tightly coupled problems in biological informatics or data science that addresses a biological research problem that can include but is not limited to: novel databases, including standards, ontologies, architectures, and interoperability; algorithms or software; tools for data analysis and integrating across biological scales; or scientific workflows. However, projects that develop tools for computer and information science, math or statistics, with no clear application to biology will be returned without review. Projects are expected to have a significant application to one or more biological science questions and have the potential to be used by a community of researchers beyond a single research team.
Program contacts
Awards made through this program, organization(s).
- Directorate for Biological Sciences (BIO)
- Division of Biological Infrastructure (BIO/DBI)
- Search Menu
- Author Guidelines
- Submission Site
- Open Access
- About Briefings in Bioinformatics
- Journals Career Network
- Editorial Board
- Advertising and Corporate Services
- Self-Archiving Policy
- Journals on Oxford Academic
- Books on Oxford Academic
Proposals for Special Issues
Briefings in bioinformatics welcomes proposals for special themed issues.
Special issues normally comprise eight to ten articles of up to 5,000 words each. Suggestions or proposals for special themed issues should be addressed to the editorial office ( [email protected] ) in the first instance.
Previous themed issues have included:
- Collaborative Bioinformatics and RNA Analysis
- Orthology and Applications
- Computational Methods for Drug Repurposing
- Validation in Bioinformatics and Molecular Medicine
- Education in Bioinformatics
- Second Generation Sequencing
- Parallel and Ubiquitous Methods and Tools in Systems Biology
- Current progress in Bioinformatics 2010
- Plant Genomics
- Challenges in Bioinformatics and Computational Biology
- Semantic Web for Health Care and Life Sciences: A Review of the State of the Art
- Database Integration in Life Sciences
- Critical Technologies for Bioinformatics
- Computational Proteomics
- Current Progress in Bioinformatics: 2007
- Integrative Biology
- Knowledge Integration and Web Communities
- Recommend to your Library
Affiliations
- Online ISSN 1477-4054
- Copyright © 2024 Oxford University Press
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
Advertisement
Bioinformatics Approaches to Stem Cell Research
- Bioinformatics and Stem Cell (R Hart, Section Editor)
- Published: 31 May 2018
- Volume 4 , pages 314–325, ( 2018 )
Cite this article
- Jia Zhou 1 &
- Renee L. Sears 1
840 Accesses
3 Citations
Explore all metrics
Purpose of Review
This review article provides an overview of the bioinformatics frameworks developed in recent years that will facilitate analysis and promote the application of stem cells to medical research.
Recent Findings
High-throughput profiling techniques at the transcript, epigenetic, and proteomic level have uncovered unique molecular signatures of stem cells that underlie their powerful capacity. A central question in stem cell research has been: “to what extent do ‘induced’ cells resemble their in vivo counterparts?” Although studies have shown overall similarity between in vitro engineered cells and their in vivo counterparts, significant deviations that lead to functional aberrations have been identified, both between pluripotent stem cell types (e.g., ESCs vs. iPSCs) and between in vitro differentiated specialized cells and their in vivo counterparts (e.g., induced pancreatic cells vs. in vivo pancreatic cells). A number of bioinformatics approaches have emerged during the past several decades, either for classification of multiple stem cell lines based on their overall molecular patterns, or for further in-depth exploration and identification of regulatory markers that can be used for differentiating cell lines or for lineage conversions.
Advancements in bioinformatics approaches in stem cell research will enhance our ability to define molecular signatures of stem cells and will accelerate the application of these stem cells to regenerative medicine.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions
Similar content being viewed by others

Transcriptomics and proteomics in stem cell research
Hai Wang, Qian Zhang & Xiangdong Fang
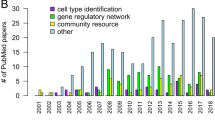
Stems cells, big data and compendium-based analyses for identifying cell types, signalling pathways and gene regulatory networks
Md Humayun Kabir & Michael D. O’Connor
Systems-Based Technologies in Profiling the Stem Cell Molecular Framework for Cardioregenerative Medicine
Saranya P. Wyles, Randolph S. Faustino, … Timothy J. Nelson
Mitalipov S, Wolf D. Totipotency, pluripotency and nuclear reprogramming. Adv Biochem Eng Biotechnol. 2009;114:185–99.
PubMed PubMed Central CAS Google Scholar
Birbrair A, Frenette PS. Niche heterogeneity in the bone marrow. Ann N Y Acad Sci. 2016;1370(1):82–96.
Article PubMed PubMed Central Google Scholar
Seale P, Asakura A, Rudnicki MA. The potential of muscle stem cells. Dev Cell. 2001;1(3):333–42.
Article PubMed CAS Google Scholar
Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78(12):7634–8.
Article PubMed PubMed Central CAS Google Scholar
Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292(5819):154–6.
Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–7.
Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76.
Eiges R, Urbach A, Malcov M, Frumkin T, Schwartz T, Amit A, et al. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell. 2007;1(5):568–77.
Ebert AD, Yu J, Rose FF, Mattis VB, Lorson CL, Thomson JA, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457(7227):277–80.
Liang P, Lan F, Lee AS, Gong T, Sanchez-Freire V, Wang Y, et al. Drug screening using a library of human induced pluripotent stem cell-derived cardiomyocytes reveals disease-specific patterns of cardiotoxicity. Circulation. 2013;127(16):1677–91.
Zhao Q, Wang X, Wang S, Song Z, Wang J, Ma J. Cardiotoxicity evaluation using human embryonic stem cells and induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Res Ther. 2017;8(1):54.
Perin L, Giuliani S, Sedrakyan S, da Sacco S, de Filippo RE. Stem cell and regenerative science applications in the development of bioengineering of renal tissue. Pediatr Res. 2008;63(5):467–71.
Article PubMed Google Scholar
Wong SS, Bernstein HS. Cardiac regeneration using human embryonic stem cells: producing cells for future therapy. Regen Med. 2010;5(5):763–75.
Zaret KS, Grompe M. Generation and regeneration of cells of the liver and pancreas. Science. 2008;322(5907):1490–4.
Dubois NC, Craft AM, Sharma P, Elliott DA, Stanley EG, Elefanty AG, et al. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat Biotechnol. 2011;29(11):1011–8.
Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453(7194):524–8.
Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25(9):1015–24.
Shiba Y, Fernandes S, Zhu WZ, Filice D, Muskheli V, Kim J, et al. Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature. 2012;489(7415):322–5.
D’Amour KA, Bang AG, Eliazer S, Kelly OG, Agulnick AD, Smart NG, et al. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24(11):1392–401.
Nistor GI, Totoiu MO, Haque N, Carpenter MK, Keirstead HS. Human embryonic stem cells differentiate into oligodendrocytes in high purity and myelinate after spinal cord transplantation. Glia. 2005;49(3):385–96.
Yan Y, Yang D, Zarnowska ED, du Z, Werbel B, Valliere C, et al. Directed differentiation of dopaminergic neuronal subtypes from human embryonic stem cells. Stem Cells. 2005;23(6):781–90.
Reubinoff BE, Itsykson P, Turetsky T, Pera MF, Reinhartz E, Itzik A, et al. Neural progenitors from human embryonic stem cells. Nat Biotechnol. 2001;19(12):1134–40.
Perrier AL, Tabar V, Barberi T, Rubio ME, Bruses J, Topf N, et al. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci U S A. 2004;101(34):12543–8.
Bilic J, Izpisua Belmonte JC. Concise review: induced pluripotent stem cells versus embryonic stem cells: close enough or yet too far apart? Stem Cells. 2012;30(1):33–41.
Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298(5593):597–600.
Sperger JM, Chen X, Draper JS, Antosiewicz JE, Chon CH, Jones SB, et al. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci U S A. 2003;100(23):13350–5.
Bhattacharya B, Miura T, Brandenberger R, Mejido J, Luo Y, Yang AX, et al. Gene expression in human embryonic stem cell lines: unique molecular signature. Blood. 2004;103(8):2956–64.
Mathur D, Danford TW, Boyer LA, Young RA, Gifford DK, Jaenisch R. Analysis of the mouse embryonic stem cell regulatory networks obtained by ChIP-chip and ChIP-PET. Genome Biol. 2008;9(8):R126.
Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38(4):431–40.
Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S, et al. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28(4):713–20.
Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–26.
Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–43.
Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1(3):299–312.
Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1(3):286–98.
Li D, Zhang B, Xing X, Wang T. Combining MeDIP-seq and MRE-seq to investigate genome-wide CpG methylation. Methods. 2015;72:29–40.
Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28(10):1097–105.
Elliott G, Hong C, Xing X, Zhou X, Li D, Coarfa C, et al. Intermediate DNA methylation is a conserved signature of genome regulation. Nat Commun. 2015;6:6363.
Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5(1):111–23.
Hu BY, Weick JP, Yu J, Ma LX, Zhang XQ, Thomson JA, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A. 2010;107(9):4335–40.
Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41(12):1350–3.
Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27(4):353–60.
Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471(7336):68–73.
Ghosh Z, Wilson KD, Wu Y, Hu S, Quertermous T, Wu JC. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS One. 2010;5(2):e8975.
Marchetto MC, et al. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS One. 2009;4(9):e7076.
Bar-Nur O, Russ HA, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9(1):17–23.
Manandhar D, Song L, Kabadi A, Kwon JB, Edsall LE, Ehrlich M, et al. Incomplete MyoD-induced transdifferentiation is associated with chromatin remodeling deficiencies. Nucleic Acids Res. 2017;45(20):11684–99.
Bian Q, Cahan P. Computational tools for stem cell biology. Trends Biotechnol. 2016;34(12):993–1009.
Bastanlar Y, Ozuysal M. Introduction to machine learning. Methods Mol Biol. 2014;1107:105–28.
Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20(3):273–97.
Google Scholar
Xu H, Lemischka IR, Ma'ayan A. SVM classifier to predict genes important for self-renewal and pluripotency of mouse embryonic stem cells. BMC Syst Biol. 2010;4:173.
Scheubert L, Schmidt R, Repsilber D, Lustrek M, Fuellen G. Learning biomarkers of pluripotent stem cells in mouse. DNA Res. 2011;18(4):233–51.
Jensen FV. An introduction to Bayesian networks. New York: Springer; 1996. 178 p
Woolf PJ, Prudhomme W, Daheron L, Daley GQ, Lauffenburger DA. Bayesian analysis of signaling networks governing embryonic stem cell fate decisions. Bioinformatics. 2005;21(6):741–53.
Dowell KG, Simons AK, Wang ZZ, Yun K, Hibbs MA. Cell-type-specific predictive network yields novel insights into mouse embryonic stem cell self-renewal and cell fate. PLoS One. 2013;8(2):e56810.
Makhoul J. Artificial neural networks. Investig Radiol. 1990;25(6):748–50.
Article CAS Google Scholar
Bidaut G, Stoeckert CJ Jr. Characterization of unknown adult stem cell samples by large scale data integration and artificial neural networks. Pac Symp Biocomput. 2009:356–67.
Jain AK, Murty MN, Flynn PJ. Data clustering: a review. ACM Comput Surv. 1999;31(3):264–323.
Article Google Scholar
Chin MH, Pellegrini M, Plath K, Lowry WE. Molecular analyses of human induced pluripotent stem cells and embryonic stem cells. Cell Stem Cell. 2010;7(2):263–9.
Pearson K. On lines and planes of closest fit to systems of points in space. Philos Mag. 1901;2(7–12):559–72.
Ulloa-Montoya F, Kidder BL, Pauwelyn KA, Chase LG, Luttun A, Crabbe A, et al. Comparative transcriptome analysis of embryonic and adult stem cells with extended and limited differentiation capacity. Genome Biol. 2007;8(8):R163.
Van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2570–605.
Bao S, Tang WWC, Wu B, Kim S, Li J, Li L, et al. Derivation of hypermethylated pluripotent embryonic stem cells with high potency. Cell Res. 2018;28(1):22–34.
Rizvi AH, Camara PG, Kandror EK, Roberts TJ, Schieren I, Maniatis T, et al. Single-cell topological RNA-seq analysis reveals insights into cellular differentiation and development. Nat Biotechnol. 2017;35(6):551–60.
Baum LE, Eagon JA. An inequality with applications to statistical estimation for probabilistic functions of Markov processes and to a model for ecology. Bull Am Math Soc. 1967; 73 (3):360.
Baum LE, Petrie T. Statistical inference for probabilistic functions of finite state Markov chains. Ann Math Stat. 1966; 37 (6):1554.
Larson JL, Yuan GC. Epigenetic domains found in mouse embryonic stem cells via a hidden Markov model. BMC Bioinformatics. 2010;11:557.
Davidson EH. The regulatory genome : gene regulatory networks in development and evolution. Burlington: Academic; 2006. 289 p
Zhou Q, Chipperfield H, Melton DA, Wong WH. A gene regulatory network in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104(42):16438–43.
Pimanda JE, Gottgens B. Gene regulatory networks governing haematopoietic stem cell development and identity. Int J Dev Biol. 2010;54(6–7):1201–11.
Swiers G, Patient R, Loose M. Genetic regulatory networks programming hematopoietic stem cells and erythroid lineage specification. Dev Biol. 2006;294(2):525–40.
Ouyang Z, Zhou Q, Wong WH. ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc Natl Acad Sci U S A. 2009;106(51):21521–6.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50.
Faustino RS, Behfar A, Perez-Terzic C, Terzic A. Genomic chart guiding embryonic stem cell cardiopoiesis. Genome Biol. 2008;9(1):R6.
Palmer NP, Schmid PR, Berger B, Kohane IS. A gene expression profile of stem cell pluripotentiality and differentiation is conserved across diverse solid and hematopoietic cancers. Genome Biol. 2012;13(8):R71.
Cahan P, Li H, Morris SA, Lummertz da Rocha E, Daley GQ, Collins JJ. CellNet: network biology applied to stem cell engineering. Cell. 2014;158(4):903–15.
Roost MS, van Iperen L, Ariyurek Y, Buermans HP, Arindrarto W, Devalla HD, et al. KeyGenes, a tool to probe tissue differentiation using a human fetal transcriptional atlas. Stem Cell Reports. 2015;4(6):1112–24.
Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, et al. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144(3):439–52.
Rackham OJ, et al. A predictive computational framework for direct reprogramming between human cell types. Nat Genet. 2016;48(3):331–5.
Zhou X, Meng G, Nardini C, Mei H. Systemic evaluation of cellular reprogramming processes exploiting a novel R-tool: eegc. Bioinformatics. 2017;33(16):2532–8.
Yamamizu K, Piao Y, Sharov AA, Zsiros V, Yu H, Nakazawa K, et al. Identification of transcription factors for lineage-specific ESC differentiation. Stem Cell Reports. 2013;1(6):545–59.
Warsow G, Greber B, Falk SSI, Harder C, Siatkowski M, Schordan S, et al. ExprEssence—revealing the essence of differential experimental data in the context of an interaction/regulation net-work. BMC Syst Biol. 2010;4:164.
Cinghu S, Yellaboina S, Freudenberg JM, Ghosh S, Zheng X, Oldfield AJ, et al. Integrative framework for identification of key cell identity genes uncovers determinants of ES cell identity and homeostasis. Proc Natl Acad Sci U S A. 2014;111(16):E1581–90.
Pinto JP, Kalathur RK, Oliveira DV, Barata T, Machado RSR, Machado S, et al. StemChecker: a web-based tool to discover and explore stemness signatures in gene sets. Nucleic Acids Res. 2015;43(W1):W72–7.
Sandler VM, Lis R, Liu Y, Kedem A, James D, Elemento O, et al. Reprogramming human endothelial cells to haematopoietic cells requires vascular induction. Nature. 2014;511(7509):312–8.
Correa-Cerro LS, Piao Y, Sharov AA, Nishiyama A, Cadet JS, Yu H, et al. Generation of mouse ES cell lines engineered for the forced induction of transcription factors. Sci Rep. 2011;1:167.
Nishiyama A, Xin L, Sharov AA, Thomas M, Mowrer G, Meyers E, et al. Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell Stem Cell. 2009;5(4):420–33.
Ernst M, Dawud RA, Kurtz A, Schotta G, Taher L, Fuellen G. Comparative computational analysis of pluripotency in human and mouse stem cells. Sci Rep. 2015;5:7927.
Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573(1–3):83–92.
Wells CA, Mosbergen R, Korn O, Choi J, Seidenman N, Matigian NA, et al. Stemformatics: visualisation and sharing of stem cell gene expression. Stem Cell Res. 2013;10(3):387–95.
Sandie R, Palidwor GA, Huska MR, Porter CJ, Krzyzanowski PM, Muro EM, et al. Recent developments in StemBase: a tool to study gene expression in human and murine stem cells. BMC Res Notes. 2009;2:39.
Yu J, Xing X, Zeng L, Sun J, Li W, Sun H, et al. SyStemCell: a database populated with multiple levels of experimental data from stem cell differentiation research. PLoS One. 2012;7(7):e35230.
Mallon BS, Chenoweth JG, Johnson KR, Hamilton RS, Tesar PJ, Yavatkar AS, et al. StemCellDB: the human pluripotent stem cell database at the National Institutes of Health. Stem Cell Res. 2013;10(1):57–66.
Bagger FO, Rapin N, Theilgaard-Mönch K, Kaczkowski B, Thoren LA, Jendholm J, et al. HemaExplorer: a database of mRNA expression profiles in normal and malignant haematopoiesis. Nucleic Acids Res. 2013;41(Database issue):D1034–9.
Bagger FO, Sasivarevic D, Sohi SH, Laursen LG, Pundhir S, Sønderby CK, et al. BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res. 2016;44(D1):D917–24.
Som A, Harder C, Greber B, Siatkowski M, Paudel Y, Warsow G, et al. The PluriNetWork: an electronic representation of the network underlying pluripotency in mouse, and its applications. PLoS One. 2010;5(12):e15165.
Xu H, et al. ESCAPE: database for integrating high-content published data collected from human and mouse embryonic stem cells. Database (Oxford). 2013;2013:bat045.
Pinto JP, Machado RSR, Magno R, Oliveira DV, Machado S, Andrade RP, et al. StemMapper: a curated gene expression database for stem cell lineage analysis. Nucleic Acids Res. 2018;46(D1):D788–93.
Clancy JL, Patel HR, Hussein SMI, Tonge PD, Cloonan N, Corso AJ, et al. Small RNA changes en route to distinct cellular states of induced pluripotency. Nat Commun. 2014;5:5522.
Hussein SM, et al. Genome-wide characterization of the routes to pluripotency. Nature. 2014;516(7530):198–206.
Lee DS, Shin JY, Tonge PD, Puri MC, Lee S, Park H, et al. An epigenomic roadmap to induced pluripotency reveals DNA methylation as a reprogramming modulator. Nat Commun. 2014;5:5619.
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41(Database issue):D991–5.
PubMed CAS Google Scholar
Karnik R, Meissner A. Browsing (Epi)genomes: a guide to data resources and epigenome browsers for stem cell researchers. Cell Stem Cell. 2013;13(1):14–21.
Zhou X, Maricque B, Xie M, Li D, Sundaram V, Martin EA, et al. The human epigenome browser at Washington University. Nat Methods. 2011;8(12):989–90.
Zhou X, Lowdon RF, Li D, Lawson HA, Madden PAF, Costello JF, et al. Exploring long-range genome interactions using the WashU epigenome browser. Nat Methods. 2013;10(5):375–6.
Hemphill EE, Dharia AP, Lee C, Jakuba CM, Gibson JD, Kolling FW, et al. SCLD: a stem cell lineage database for the annotation of cell types and developmental lineages. Nucleic Acids Res. 2011;39(Database issue):D525–33.
Jung M, Peterson H, Chavez L, Kahlem P, Lehrach H, Vilo J, et al. A data integration approach to mapping OCT4 gene regulatory networks operative in embryonic stem cells and embryonal carcinoma cells. PLoS One. 2010;5(5):e10709.
Schulz H, Kolde R, Adler P, Aksoy I, Anastassiadis K, Bader M, et al. The FunGenES database: a genomics resource for mouse embryonic stem cell differentiation. PLoS One. 2009;4(9):e6804.
Watkins NA, Gusnanto A, de Bono B, De S, Miranda-Saavedra D, Hardie DL, et al. A HaemAtlas: characterizing gene expression in differentiated human blood cells. Blood. 2009;113(19):e1–9.
Download references
Acknowledgments
We thank Ting Wang for valuable suggestions on the manuscript and Paul Dillingham for editorial contribution.
Author information
Authors and affiliations.
Department of Genetics, Center for Genome Sciences and Systems Biology, Washington University School of Medicine, St. Louis, MO, 63108, USA
Jia Zhou & Renee L. Sears
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Jia Zhou .
Ethics declarations
Conflict of interest.
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Bioinformatics and Stem Cell
Rights and permissions
Reprints and permissions
About this article
Zhou, J., Sears, R.L. Bioinformatics Approaches to Stem Cell Research. Curr Pharmacol Rep 4 , 314–325 (2018). https://doi.org/10.1007/s40495-018-0143-4
Download citation
Published : 31 May 2018
Issue Date : August 2018
DOI : https://doi.org/10.1007/s40495-018-0143-4
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Bioinformatics
- Find a journal
- Publish with us
- Track your research

Grant Proposal Assistance
- CBCB Bioinformatics Data Science Core
Writing a Grant or Contract Proposal?
The Biomedical and Life Sciences have become increasingly data intensive, and success of a proposal can hinge upon a solid plan and suitable expertise for data analysis.
We are happy to provide you with assistance on the bioinformatic and data science aspects of your proposal. We are happy to arrange a free consultation to discuss the needs of your proposed project and to provide support, including:
- experimental design
- relevant bioinformatics methods
- budget planning
- facilities and resources statements
- letters of support
In addition, our experts may be available to act as project participants providing an additional level of expertise to your proposal. Grant proposal support is provided free of charge.

An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- J Med Internet Res
- PMC10407648

Ten Topics to Get Started in Medical Informatics Research
Markus wolfien.
1 Institute for Medical Informatics and Biometry, Faculty of Medicine Carl Gustav Carus, Technische Universität Dresden, Dresden, Germany
2 Center for Scalable Data Analytics and Artificial Intelligence, Dresden, Germany
Najia Ahmadi
3 Core Unit Data Integration Center, University Medicine Greifswald, Greifswald, Germany
Sophia Grummt
Kilian-ludwig heine, dagmar krefting.
4 Department of Medical Informatics, University Medical Center, Goettingen, Germany
Andreas Kühn
Ines reinecke, julia scheel.
5 Department of Systems Biology and Bioinformatics, University of Rostock, Rostock, Germany
Tobias Schmidt
6 Institute for Medical Informatics, University of Applied Sciences Mannheim, Mannheim, Germany
Paul Schmücker
Christina schüttler.
7 Central Biobank Erlangen, University Hospital Erlangen, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany

Dagmar Waltemath
8 Department of Medical Informatics, University Medicine Greifswald, Greifswald, Germany
Michele Zoch
Martin sedlmayr.
The vast and heterogeneous data being constantly generated in clinics can provide great wealth for patients and research alike. The quickly evolving field of medical informatics research has contributed numerous concepts, algorithms, and standards to facilitate this development. However, these difficult relationships, complex terminologies, and multiple implementations can present obstacles for people who want to get active in the field. With a particular focus on medical informatics research conducted in Germany, we present in our Viewpoint a set of 10 important topics to improve the overall interdisciplinary communication between different stakeholders (eg, physicians, computational experts, experimentalists, students, patient representatives). This may lower the barriers to entry and offer a starting point for collaborations at different levels. The suggested topics are briefly introduced, then general best practice guidance is given, and further resources for in-depth reading or hands-on tutorials are recommended. In addition, the topics are set to cover current aspects and open research gaps of the medical informatics domain, including data regulations and concepts; data harmonization and processing; and data evaluation, visualization, and dissemination. In addition, we give an example on how these topics can be integrated in a medical informatics curriculum for higher education. By recognizing these topics, readers will be able to (1) set clinical and research data into the context of medical informatics, understanding what is possible to achieve with data or how data should be handled in terms of data privacy and storage; (2) distinguish current interoperability standards and obtain first insights into the processes leading to effective data transfer and analysis; and (3) value the use of newly developed technical approaches to utilize the full potential of clinical data.
Introduction
Digital health care information, as opposed to analog information, empowers clinicians, researchers, and patients with a wealth of information aiming to improve diagnosis, therapy outcome, and clinical care in general. According to Wyatt and Liu [ 1 ], medical informatics is the study and application of methods to improve the management of patient data, clinical knowledge, population data, and other information relevant to patient care and community health. Medical informatics can be seen as the subset of health informatics that is focused on clinical care, while the latter encompasses a wider range of applications. However, knowing, integrating, and using current computational technologies bears numerous pitfalls, limitations, and questions [ 2 ]. To shed light on current standards, applications, and underlying technologies, we present 10 topics to get started in the field of medical informatics research. Our key objective here was to improve interdisciplinary communication among stakeholders (eg, clinicians, experimental researchers, computer scientists, students, patient representatives), thereby bringing everyone on the same page of state-of-the-art medical informatics practices. In particular, improved interdisciplinary communication is essential in real-world problems and can be motivated by the following aspects:
- Advancing open research: Open collaboration between parties from different disciplines can lead to new research questions, innovative approaches, and novel discoveries [ 3 ].
- Bridging knowledge domains: Interdisciplinary communication can stimulate novel solutions, allowing researchers to gain a more comprehensive understanding of a specific problem or phenomenon [ 4 ], or can improve clinical decision-making [ 5 ].
- Addressing complex problems: Complex problems, such as the latest disease outbreak, require input from multiple domains to be comprehensively understood. Here, interdisciplinary communication is one key aspect to pinpoint the root causes and develop effective solutions [ 6 ].
- Promoting scientific inclusivity and diversity: Interdisciplinary communication was recently shown to foster diversity and inclusivity in science, by bringing together researchers from different backgrounds, cultures, and perspectives [ 7 , 8 ].
Here, we describe in detail how the initial topics have been selected from the literature and what design principles and structure each topic follows. A brief outline of the utilized methods for topic dissemination and an exemplary embedding into an educational training program are also presented.
Topic Selection
The initial topics were defined based on current developments in the health informatics field and an increasing number of published manuscripts between 2000 and 2021 (based on title-abstract-keyword screening in Scopus using the keywords “Health” AND “Informatics” AND “domain”) in the respective subdomains ( Figure 1 A). After a first definition of the specific topics, these were critically revised by internal and external domain experts, as well as scientists previously not familiar with medical informatics research.

Schematic summary and representation of the presented topics: (A) brief literature screening (title-abstract-keywords) for published manuscripts between 2000 and 2021, and the y-axis gap provides improved visibility of the less-occurring keywords; (B) most common topic terminologies, keywords (color-coded sections), and potential connections (grey) among topics in the medical informatics research domain. CDSS: clinical decision support system; CIS: clinical information system; EHR: electronic health record; ETL: extract, transform, and load; FAIR: findable, accessible, interoperable, reusable; FHIR: Fast Healthcare Interoperability Resources; GDPR: General Data Protection Regulation; i2b2: Informatics for Integrating Biology and the Bedside; OMOP: Observational Medical Outcomes Partnership.
Topic Design
The initial number of important topics and keywords exceeded the anticipated number of 10 topics, which found inspiration from the “Ten Simple Rules” collection in PLOS Computational Biology [ 9 ]. This is why the authors merged the most matching terms topic wise into groups. These groups finally produced topics that represent the broad range of the medical informatics domain in 3 main concepts, namely “Regulations and concepts,” “Harmonization and processing,” and “Evaluation, visualization, and dissemination” ( Figure 1 B). Figure 1 B also shows the initial keywords for each individual topic, as well as potential cross references between topics, which are highlighted in grey. The following sections provide important “do's and don'ts,” practical hints, and best practice guidelines. Further in-depth resources and practical tutorials will provide basic introductions to the referred domains. Kohane et al [ 10 ] already showed the importance of such clarifying introductions. This work extends the initial study and, in addition, provides detailed examples from the German national Medical Informatics Initiative (MII) [ 11 ].
All topics were divided into 3 parts to improve comprehension by the readers:
- Introduction: Background definitions for the specific context that motivated the topic
- Insight: Practical context to get started, including how to avoid pitfalls, state current limitations, and address current challenges
- Impact: Take home message and useful resources and best practices to deepen knowledge about the topic
Topic Utilization, Extension, and Embedding
Since it is of the utmost importance to keep the content current and as versatile as possible, we initiated an online resource at GitHub, in which contributions are highly emphasized [ 12 ]. Here, keywords and the corresponding literature are collected to allow for swift extension of the currently presented literature body in this article. In addition, the introduction of novel important topics that are not covered in this article might be included. To additionally demonstrate the practicability and adaptability of our proposed topic content, we exemplarily present how these can be embedded in higher education training and share external, introductory hands-on material ( Table 1 ).
Summary of tutorials and hands-on material about medical informatics standards and applications.
a SNOMED CT: Systematized Nomenclature of Medicine and Clinical Terms.
b ETL: extract, transform, and load.
c OMOP: Observational Medical Outcomes Partnership.
d CDM: common data model.
e FHIR: Fast Healthcare Interoperability Resources.
f OHDSI: Observational Health Data Sciences and Informatics.
g PLP: patient-level prediction.
h ODI: Open Data Institute.
Regulations and Concepts
Topic 1: privacy and ethics—“data privacy and ethics are the most important assets in the clinical domain.”.
Health information is sensitive and hence needs to be highly protected and should not be generously shared. Sharing regulations and data privacy matters are defined in the European General Data Protection Regulation (GDPR) [ 13 ]. The implementation of the GDPR is an ongoing process as the quickly evolving technology, data, and scientific practices demand continuous improvement, which include periodic adaptations of the technical and legal aspects [ 14 , 15 ]. In terms of ethics and with the rise of novel technologies, like artificial intelligence (AI), the possible re-identification of data, such as images and genomic information, is a major concern [ 16 , 17 ].
Anonymization is one important way to keep data private. It can also be achieved for high-dimensional data by changing patient-specific identifiers through removal, substitution, distortion, generalization, or aggregation [ 18 ]. In contrast, data pseudonymization is another de-identification procedure by which personally identifiable information fields within a data record are replaced by one or more artificial identifiers or pseudonyms [ 19 ]. To overcome the paucity of annotated medical data in real-world settings and (fully) save the patients’ anonymity, synthetic data generation is used to increase the diversity in data sets and to enhance the robustness and adaptability of AI models [ 20 ]. To conform with ethical regulations in a research context, medical data are only available in a highly controlled manner and according to strict procedures. New concepts, such as “systemic oversight” [ 21 ] or “embedded ethics” [ 22 ], might be needed to tackle the new data-driven developments around “medical big data” and AI in health care. To engage with the adoption of broad consent, systemic oversight was suggested as an approach, in which mechanisms like auditing mechanisms, expert advice, and public engagement initiatives (among others) should be adapted as additional layers to the newly arising ecosystem of health data [ 21 ]. Recently, embedded ethics was jointly suggested by ethicists and developers to address ethical issues via an iterative and continuous process from the outset of development, which could be an effective means of integrating robust ethical considerations into practical development [ 22 ]. A digital representation of information encoded in signed consent forms is needed to facilitate common data use and sharing, as already implemented in an MII informed consent template [ 23 ].
As a researcher in medical informatics, it is inevitable to be informed and knowledgeable about the fact that patients own their medical records and any use of those data requires great care. In Germany, health care providers can only use the data for first medical use. Secondary use, like research, needs to be approved by either broad or individual consent, which can be made available via the electronic health record (EHR). In addition to digitization efforts, it is still a considerable hurdle to convince patients to make their data available for medical research because personal skepticism commonly makes the entire data acquisition process more difficult [ 24 ]. Here, well-received external communication, transparency, and increased awareness are necessary for substantial improvements. In general, it is a balance between privacy, patient needs, and the use of data for the common good versus economic interests [ 25 ]. In particular, one should be aware of the specific legal regulations that apply within the country and additionally get in touch with the relevant data protection departments. Following this, a plan for infrastructure that meets these regulations and that contains, for example, a trustee for the electronic recording of patient consent and anonymization or direct pseudonymization processes to collect the data needs to be developed. Risk assessments for potential data leakage, approvals by ethics committee, as well as consultation with a data protection officer are essential considerations to further assure data security.
Topic 2: EHR and Clinical Information Systems—“Get to Know Your Clinical Information System to Understand the Required Data.”
Hospitals run clinical information systems (CIS) to collect, store, and alter clinical data about patients. A CIS, independent of the specialization and specific vendor, covers many clinical subdomains and integrates patient-related data to support doctors in their daily routine. Without a doubt, medical data are only useful if meaningful information can be derived from them. This requires high-quality data sets, seamless communication across IT systems, and standard data formats that can be processed by humans and machines [ 2 ]. Typical challenges in clinical IT implementations, especially for patient recruitment systems, were recently evaluated by Fitzer et al [ 26 ] for 10 German university hospitals, including requirements for data, infrastructure, and workflow integration. The implementation of an EHR, including an individual's medical data in a bundled form, into the CIS is a key aspect to prevent low reliability and poor user-friendliness of EHRs, which has recently been shown to affect time pressure among medical staff [ 27 ]. For example, in Scandinavia, the United States, and the United Kingdom, the Open Notes initiative [ 28 ] facilitates patients’ access to EHRs and health data sharing via “PatientsKnowBest ” to give health care professionals and families direct access to medical information [ 29 ].
An EHR is used primarily for the purposes of setting objectives and planning patient care, documenting the delivery of care, and assessing the outcomes of care [ 30 ]. EHRs have so far consisted of unstructured, narrative text as well as structured, coded data. Thus, it will be necessary to implement more systematic terminologies and codes so that the data contained in these records can be reused in clinical research, health care management, health services planning, and government reporting in an improved manner [ 31 , 32 ]. Since the domain of medical informatics is rather new, there are many possibilities for software solutions to improve EHR-related issues [ 33 ]. Exemplary for the EHR domain, the Systematized Nomenclature of Medicine and Clinical Terms (SNOMED CT) is utilized to develop comprehensive high-quality clinical content [ 34 ]. It provides a standardized way to represent clinical phrases captured by the clinician and enables automatic interpretation of these, which is showcased in a “five-step briefing” [ 35 ]. Interestingly, the number of annual publications on this subject has decreased since 2012. However, the need for a formal semantic representation of free text in health care remains, and automatic encoding into a compositional ontology could be a solution [ 36 ]. In terms of usability and user acceptance, evaluations and improvements of EHRs and clinical decision support systems (CDSS) are currently ongoing [ 37 ], for which already well-received examples can be attributed to CeoSYS [ 38 ] or the IPSS-M Risk Calculator [ 39 ]. Moreover, the actions of patients directly contributing to their own EHR records are also being evaluated. The study by Klein et al [ 40 ] indicates that such an approach facilitates the development of individual solutions for each patient, which in turn requires a flexible EHR during the course of a treatment process. Additionally, it was argued that data incorporation via different devices can also facilitate the convenient utilization of the application and, hence, may increase secondary use.
Modern CIS support the interaction by doctors and patients with the recorded patient data (eg, using the EHR or patient portals, eHealth platforms). It is important to understand the basic architecture, especially challenges [ 26 ], of the hospital IT infrastructure to know where data are located and how they can be retrieved and integrated. Major improvements can be made when supporting international standards for data exchange. Beyond standard EHR, this includes interoperability standards like Fast Healthcare Interoperability Resources (FHIR; see Topic 6) and standard data models like the Observational Medical Outcomes Partnership (OMOP; see Topic 7). These criteria should be considered with every new order of clinical systems.
Topic 3: Data Provenance—“Trace Your Data, Even Within Large-scale Efforts.”
Meaningful and standardized metadata facilitate the interpretation of, retrieval of, and access to data [ 41 ]. When explainable data are processed with interoperable tools, scientists can create automated and reusable workflows and provide access to reproducible research outcomes and data analysis pipelines [ 42 ].
Data provenance describes the history of digital objects, where they came from, how they came to be in their present state, and who or what acted upon them [ 43 ]. In health care, provenance maintains the integrity of digital objects (eg, the results of data analyses engender greater trust if their provenance shows how they were obtained). In addition, it can be used to deliver auditability and transparency, specifically, in learning health systems, and it is applicable across a range of applications [ 44 ]. Inau et al [ 45 ] argued that the lessons learned from “FAIRification” processes in other domains will also support evidence-based clinical practice and research transparency in the era of big medical data and open research. Further work demonstrated that a findable, accessible, interoperable, reusable (FAIR) research data management plan can provide a data infrastructure in the hospital for machine-actionable digital objects [ 46 ]. Recently, the openEHR approach was also suggested for creating FAIR-compliant clinical data repositories as an alternative representation [ 47 ].
Key data management requirements are defined by the FAIR guiding principles [ 48 ]. Since data protection laws led to additional requirements for data privacy and data security, the FAIR-Health principles focused on defining additional requirements for information on the sample material used from biobanks, for provenance information, and incentive schemes [ 49 ]. Further work is needed to establish provenance frameworks in health research infrastructures [ 50 ].
Topic 4: Data Sharing—“If Data Won’t Come to the Model, the Model Must Go to the Data.”
Cross-sectional medical data-sharing is critical in modern clinical practice and medical research, in which the challenge of privacy-preserving transfer and utility needs to be addressed [ 51 ]. In order to facilitate high reuse of the data, a decentralized computational scheme that treats the available data as part of a federated (virtual) database, avoiding centralized data collection, processing, and raw data exchanges, is still needed in many countries to analyze large and widespread clinical data [ 52 ].
One possible solution for this federated learning approach is DataSHIELD [ 53 ]. In particular, orchestrating privacy-protected analyses of “medical big data'' from different resources is applicable within R and DataSHIELD [ 54 ]. Here, the developed computerized models represent mathematical concepts or trained machine learning (ML)–based approaches to solve a specific task. In this sense, the model is applied to distributed data sets of the protected (clinical) server infrastructure, and the user only sees the model results but does not retrieve any medical records. Moreover, implementations in other programming languages (eg, Python, Julia) have been introduced in the genomic domain and beyond [ 55 ]. Further concepts, such as Personal Health Train, specifically follow the FAIR principles during distributed analyses [ 56 ]. Secure multiparty computation (SMPC) is also a viable technology for solving clinical use cases that require cross-institution data exchange and collaboration [ 57 ]. Current limitations are thought to be addressed in a stepwise manner [ 58 ] or as blockchain [ 59 ].
By using approaches for distributed analyses, researchers are able to train, test, and validate their models on large-scale real-world clinical data. In combination with standardized data formats, these 2 concepts facilitate the use of those models in clinical routine, potentially in the form of a CDSS. This provides a basis for secondary use of observational data in the context of clinical trials, which show particular potential for identifying data characteristics in small cohorts (eg, identification of the individual patient risk for rare diseases or comorbidities).
Harmonization and Processing
Topic 5: extract, transform, and load (etl)—“ etl processes are computational approaches for data harmonization and data unification.”.
Data handling in medical informatics remains a major challenge. Even though most data in medicine are available electronically, the data often lack interoperability [ 60 ]. As a first step to actually use the data, processes to extract, transform, and load (ETL) are needed to obtain harmonized data from different data systems or clinical entities. One important example, among many others, reflects the uniform representation of the date and time in a common format (eg, Year-Month-Date, not Date-Month-Year). The ETL process is therefore a crucial, individual step toward data unification in large clinical systems, which must be secure, safe, and accurate [ 61 ].
The design of an ETL process faces several challenges, including the following: (1) The ETL process should be able to process huge amounts of data at once [ 62 ]; (2) the ETL process should be repeatable—if the source data change, the ETL process needs to be rerun to process the source data (Observational Health Data Sciences and Informatics [OHDSI]) [ 63 ]; (3) expert-level anonymization methodologies might be integrated into ETL workflows whenever possible [ 61 ]; and (4) there is a need to check for loss of data and compromised data integrity. The latter was highlighted in a recent study, in which inaccurate cohort identification took place because erroneous vocabulary mappings of a common data model were used (eg, ETL programming bugs and errors not captured during the quality assurance stages) [ 64 ]. Common solutions to implement ETL processes are code-based (eg, FHIR-to-OMOP [ 65 ]) or via Pentaho Data Integration, which is one of many ETL tools. Further subsequent processing may also include loading data into research data repositories, like OMOP (see Topic 7), tranSMART, and Talend Open Studio, which is a central component of the Integrated Data Repository Toolkit [ 66 ].
Since ETL processes are at the core of data handling, all risks associated with the ETL process need to be thoroughly checked, identified, and assessed, and contingency plans to mitigate these risks should be in place [ 67 ]. Once the ETL processes are executed, the resulting data will be trusted by researchers, who heavily rely on comprehensively checked data integrity to be able to conduct their research on this basis.
Topic 6: FHIR—“Set FHIR to Gain a Communication Standard for Real-time Applications at the Device-to-Device Level.”
Interoperability levels can be divided into technical, syntactic, semantic, and organizational interoperability [ 2 ]. Semantic and syntactic interoperability can be ensured by communication exchange standards, such as the FHIR [ 68 ] standard of Health Level 7 (HL7) and medical terminologies. A suitable starting point for the basic procedures is offered by FHIR drills [ 69 ] or fire.ly [ 70 ].
FHIR is one of many communication standards but will likely change the domain of clinical IT significantly [ 71 , 72 ]. As a communication standard, FHIR harmonizes data formats coming from different CIS and enables data exchange between institutions via a RESTful approach [ 73 ]. Moreover, FHIR is used to connect devices with each other, which means, in particular, that the Integrating the Healthcare Enterprise (IHE) [ 74 ] standard has been revised to support HL7 messaging as well. In turn, IHE has been developing an open-source device tool set for home and hospital use that recently enabled device control capabilities, a capability accelerated during the COVID-19 pandemic to allow nurses and physicians to operate ventilators and infusion devices outside the contaminated patient room [ 75 ].
Utilizing FHIR in multiple applications already shows its versatile and flexible use (eg, in mobile health applications [ 76 ], electrocardiogram monitoring [ 77 ], or wearable devices and precision medicine in digital health [ 72 ]). In particular, the SMART-on-FHIR technology enables third-party app development for health care applications [ 78 ] and encompasses feasible, secure, and time- and resource-efficient solutions [ 79 , 80 ].
Topic 7: OMOP—“Use Common Data Models as Well-defined Representations of Large-scale Research Projects.”
Data harmonization enables research teams to run real-world observational studies based on heterogeneous data across country borders. Thus, harmonized data embedded in a common data model (CDM), which is an agreement about the utilization of standardized terminologies for data representation, is crucial to exchange data and results on a large scale. To foster reliability and trust in the results of observational research on real-world data, it is essential to utilize CDMs whenever possible to ensure a high degree of data analysis reproducibility.
Several CDMs exist for that purpose; the OMOP CDM from the OHDSI community is one of the most promising and established approaches. In comparison with other CDMs, such as the Sentinel CDM or Informatics for Integrating Biology and the Bedside (i2b2), the OMOP CDM has broader terminology coverage [ 81 ]. The importance of the OMOP CDM increased a lot over the last years [ 82 ], not least since the European Medicines Agency initiated the Data Analysis and Real World Interrogation Network (DARWIN) [ 83 ] project to establish a research network in Europe to gain real-world evidence based on OMOP. Moreover, representations of genomic data [ 84 ], oncology [ 85 ], and imaging projects [ 86 ] are also suitable. In addition, the common representation of the data in OMOP semantic interoperability is ensured by utilizing international terminologies and vocabularies, such as SNOMED-CT, the International Statistical Classification of Diseases and Related Health Problems (ICD), the Logical Observation Identifiers Names and Codes (LOINC), and RxNorm to represent every clinical fact in OMOP. Additionally, the open-source OHDSI software stack provides standardized methodology and libraries for data analyses (Athenahene, Atlas, HADES) and training (EHDEN Academy) [ 87 ], as well as a framework to assess and improve data quality to foster reliability and trust in research results [ 88 ].
The OMOP CDM is one possibility to represent and analyze clinical data on a research scale. Definition of new cohorts within OMOP enables researchers to quickly investigate questions spanning multiple research entities. Collectively, both FHIR and OMOP can define the structure and relations of the clinical data corpus, and the individual EHRs provide content to these standardized data reservoirs. In comparison, OMOP is commonly used for static large-scale data analysis of research data, and FHIR is more suitable for rapid data integration scenarios (ie, for real-time applications and analysis). In summary, it is important to know and utilize newly established standards to participate in broader clinical networks for research. This way, all information within the EHR is comparable across different clinical sites and research settings.
Evaluation, Visualization, and Dissemination
Topic 8: data quality—“guarantee high quality and then publish the data.”.
What is meant to be appropriate data quality for health informatics research? In this domain, data quality depends on the quality of single data elements, data completeness, data conformance, and data plausibility aspects that may considerably determine the validity and veracity of analysis results [ 89 , 90 ]. Moreover, data quality across different institutional entities and even health sectors requires additional efforts concerning the different personnel, instruments, and more [ 91 ]. High-quality data at hand is one fundamental requirement that is often difficult or impossible to achieve, which is why the generation of synthetic data can be an alternative that satisfies privacy problems as well as research needs when data are expensive, scarce, or unavailable by augmentation [ 92 ].
First, a major problem is that clinical data have to be electronically recorded, accessed, and standardized in order to run quality assessment processes [ 26 ]. In addition, it would be important to design and use the same data quality tool, standard operating procedures, or ETL mapping rules in all involved institutions. However, in real-life scenarios, there is a lack of both centrally coordinated data quality indicators and formalization of plausibility rules, as well as a repository for automatic querying of the rules, especially in ETL processes [ 93 ]. Although numerous data quality evaluation frameworks exist, no clear and widespread approach has been adopted so far [ 67 , 94 - 96 ]. Even after a well-chosen data quality procedure is properly implemented, clinical data as such cannot be published along with the performed study. As an alternative, synthetic data generation models function in the following 2 different ways: (1) The model is trained, for example, using real-world data and, once trained, will not require any data in the future (model-based approaches), and (2) the model is constantly fed with data to generate synthetic data (data-driven approaches). There are 3 different categories of algorithms used in the generation of synthetic data: probabilistic models, such as Bayesian networks [ 97 ] and Copulas [ 98 ]; ML, such as Classification and Regression Trees (CART); and deep learning methods, such as a generative adversarial network (GAN) [ 99 - 101 ] and variational autoencoder (VAE) [ 102 ].
A combination of appropriate data quality evaluation and synthetic data generation highly facilitates the development of accurate AI models, which are essential in medical studies [ 103 ]. Thus, a corpus of high-quality synthetic data with many patients can be reused by other AI experts for model development and benchmarking. Moreover, it is essential to create an infrastructure that is used across a large community of hospitals; maps the entire treatment process electronically; and only generates interoperable, structured data based on FHIR (Topic 6) and OMOP (Topic 7) in accordance with the FAIR principles (Topic 3). Afterward, one can finally run quality assessment processes.
Topic 9: Clinical Decision Support Systems—“Bring Insights, Not Additional Work, Back to the Clinics via a CDSS and Other User-Centric Applications.”
CDSS are computer systems designed to assist the medical staff with decision-making tasks about individual patients and based on clinical data [ 104 ]. The decision-making process is still, and will remain, on the shoulders of the physician [ 105 ]. The categories of CDSS include knowledge-based systems that make use of clinical rules, nonknowledge-based systems (eg, AI-based systems), and hybrid CDSS that likewise utilize clinical models and knowledge in combination with AI.
The use of a CDSS in a well-implemented clinical workflow has many positive aspects. It may lead to fewer error rates [ 106 ], accelerate rare disease diagnosis [ 107 ], increase radiologists’ job satisfaction [ 108 ], offer personalized cancer treatment [ 109 ], or help with real-time cardiovascular risk assessment [ 110 ]. Interestingly, computerized alerting systems, which are one of the most disseminated CDSS, can decrease drug-drug interactions significantly [ 111 ]. On the other hand, if done improperly, a CDSS can cause alert fatigue by creating too many alerts. If a system is not context-sensitive, alerts can even be inappropriate [ 112 ]. According to Olakotan et al [ 112 ], influencing factors of a well-designed CDSS need to include aspects about the (1) technology (eg, usability, alert presentation, workload, and data entry), (2) human (eg, training, knowledge, skills, attitude, and behavior), (3) organization (eg, rules and regulations, privacy, and security), and (4) and process (eg, waste, delay, tuning, and optimization). To avoid a lack of transparency and facilitate acceptance by physicians, especially with nonknowledge-based systems, current CDSS seek to use explainable AI approaches; however, the selection of methods used to present explanations in an informative and efficient ( clinically useful ) manner remains challenging [ 113 ]. Of note, a CDSS may also have a negative influence on the performance of physicians, especially if inadequate suggestions occur more often, which cannot be compensated with explanations [ 114 ]. However, one among many other prominent approaches to obtain such explanations via ML-based feature selection and ranking can be found in the work from Wolfien et al [ 115 ]. In terms of an OMOP-based implementation in research, there is patient-level prediction (PLP), which is designed to foster the clinical decision-making process concerning diagnoses or treatment pathways based on the EHR of the patient and the current clinical guideline. It is used to answer questions, such as identifying patients among a larger population at higher risk of a certain outcome (eg, occurrence of cancer, severe side effects, or death) by using data in standardized formats (eg, as previously described via OMOP CDM). Once the model is designed, the covariates will be extracted from the respective CDM of the target person within the cohort, and the respective outcome will be predicted (eg, via PLP [ 116 , 117 ] or other customized prediction algorithms). Importantly, the results from model prediction should first be internally validated with previously unseen data and afterward compared with established scoring systems (eg, Framingham Risk Score [ 118 ], SCORE2 [ 119 ]) to connect with already known domain-specific contexts and to prove its benefit in clinical practice. An additional validation with external data, as part of a multicenter study, can be seen as highly beneficial, in which the already presented topics of federated learning (Topic 4) and OMOP (Topic 7) could significantly foster such an essential scenario [ 120 ].
Collectively, a CDSS increases patient safety, assists in clinical management, and can be cost-effective [ 104 ]. In general, findings of even erroneous CDSS can be used to guide the design of new CDSS alerts. However, the existing risks cannot be solved solely on a technical basis and require an interdisciplinary effort. In particular, continuous, clear communication between IT professionals (developers) and health professionals (end users) during the design process is key. Only a profound understanding of the needs and requirements of either of the involved parties can lead to well-designed systems that are actually able to support and relieve physicians in doing their job.
Topic 10: Visualizations—“Improved Dissemination of Local and External Data From Computational Models by Well-defined Interactive Visualizations.”
Large volumes of data collected from patient registries, health centers, genomic databases, and public records can potentially improve the efficiency and quality of health care via enhancing the interoperability of medical systems, assisting in clinical decision-making, and delivering feedback on effective procedures [ 121 ]. However, each and every raw data point must go through different analytical processes until they become useful and interpretable at the point of care.
R and Python are 2 versatile open-source programming languages that have gained popularity for different purposes, such as preprocessing (eg, tidyverse), statistical tests (eg, dplyr), ML and deep learning (eg, mlr package, caret), visualization (eg, ggplot), and writing reports directly using knitr and R markdown (RStudio education [ 122 ]). Like R, Python offers different libraries for data science tasks (eg, open mined [ 123 ]) in addition to a library specifically for health predictive models, namely PyHealth [ 124 ]. Another versatile visualization functionality is offered for both languages via R Shiny [ 125 ] and Plotly Dash [ 126 ]. These 2 platforms enable data scientists to create interactive web applications directly from a script. The applications can be extended using embedded CSS themes, HTML widgets, and Javascript actions. There is already evidence that implementing clinical dashboards or CDSS for immediate access to current patient information can improve processes and patient outcomes [ 127 ], especially if the data sets are further evaluated and refined [ 128 ]. Similar to FHIR, OHDSI provides tools for analyzing data in the OMOP CDM, which are written in R and use Shiny for the visualization. As a plus, data already stored in the OMOP CDM format can be used in systematic studies, patient-level analysis, and population-based estimations from scratch. The cBioPortal is one prime example of a web resource for exploring, visualizing, and analyzing multidimensional data, which reduces molecular profiling data from cancer tissues and cell lines into readily understandable genetic, epigenetic, gene expression, and proteomic events [ 129 ]. It was recently demonstrated how cBioPortal can be extended and integrated with other tools to a comprehensive and easily deployable software solution that supports the work of a molecular tumor board [ 130 ] and even deliver meaningful scientific insights [ 131 ]. Another translational research platform for the construction and integration of modern clinical research charts is Informatics for i2b2, which is also at the heart of clinical research [ 132 , 133 ].
Computational approaches and data analyses are tightly connected with medical research; the visualization of such complex data for clinicians in a routine setting especially plays a larger role. The current developments of translational research platforms, such as cBioPortal and i2b2, enable swift translation of research results into the clinic, if adequately adopted and enough trained people supervise the process.
The need for qualified IT specialists in medical informatics has increased continuously in recent years and will continue to grow in the future. On the other hand, medical informatics in Germany faces problems with the promotion of young researchers. These current developments mean that vacancies in IT in hospitals and the health care industry can often not be filled or only after very long vacancies. In addition, these positions often have to be filled with nonspecialist staff due to a lack of applications. To keep track of these recent developments and provide a basis for interdisciplinary communication, we provide our list of 10 topics that could be used by different stakeholders individually ( Figure 2 ). With a particular focus in medicine, improved interdisciplinary communication has already been shown to positively impact patient outcomes and enhance employee engagement [ 134 ].

Exemplary outcome visualization of the underlying study, in which the color coding reflects the initial colors of the proposed sections; it starts with an individual perception of the term medical informatics (MI) based on the individual’s background and ends with acquisition of common domain knowledge for current important topics. CDSS: clinical decision support system; EHR: electronic health record; ETL; extract, transform, and load; FAIR; findable, accessible, interoperable, reusable; FHIR; Fast Healthcare Interoperability Resources; OMOP: Observational Medical Outcomes Partnership.
Furthermore, medical informatics has developed rapidly in recent years. This applies, for example, to new methods, techniques, tools, framework conditions, and organizational structures, especially in the field of medical data science. In particular, definitions of standards and a national digitized data corpus, namely the German Core Dataset [ 135 ], were agreed upon. The actual assessment and collection of digitized data in local university hospitals are utilized in so-called data integration centers. These interoperable research data infrastructures enable rapid multisite research, for example, with complex COVID-19 research data sets (German Corona Consensus Dataset [GECCO]) [ 136 ] including clinical data and data on biosamples from all German university hospitals in pseudonymized form (CODEX) [ 137 , 138 ] or the COVID-19 Data Portal [ 139 ]. The subsequent formation of the Network University Medicine (NUM) strengthens the existing interaction between research and patient care, stabilizes existing structures, and creates new structures that ensure more effective feedback and close cooperation between the clinics. The presented examples of NUM and CODEX, among others [ 140 ], attempt a central approach to bundle and harmonize necessary resources like broad consent or the elektronische Patientenakte (ePa), which is the implementation of EHR as a national entity to ultimately facilitate an interconnected health care system.
Finally, all those involved in medical informatics are called upon to engage in lifelong learning and continuously acquire further qualifications.
Exemplary Implementation of the Addressed Topics in the German Medical Informatics in Research and Care in University Medicine Consortium
This article offers newcomers to medical informatics a first introduction and a wealthy overview of current IT-related topics in research and patient care. Nevertheless, there is also a need for further qualification of employees through new, innovative offers for training, further education, and further training. As part of the MII [ 11 ], all consortia were asked to develop and set up appropriate offers and formats. The Medical Informatics in Research and Care in University Medicine (MIRACUM) consortium [ 141 ] has reacted and set up the part-time training and further education program “Biomedical Informatics and Data Science” [ 142 ] and introduced it at the Mannheim University of Applied Sciences in October 2020. The program includes a time-flexible and individually adaptable part-time online master’s course, as well as certificate courses and programs for further scientific education. In addition to the establishment and continuous further development of a cloud-based learning platform, many new digital and target group–oriented learning resources and application-oriented learning environments were developed and introduced for the master's program.
All 10 topics listed in this article are reflected in the curriculum of the master’s degree and have been offered and dealt with in-depth in the individual courses for more than 2 years. The demand for the master’s program and certificate courses is high, and the evaluation has shown that these topic-specific foci correspond to the training and further education needs of the target groups. One particular aspect that was not covered in the final topics refers to the underlying infrastructure needed to provide the data storage and processing backbone. This aspect would have been too technical for a more broadly set, introductory article, such as this article. A starting point for more in-depth information about this aspect can be obtained from further literature [ 143 , 144 ]. However, to offer a practical start to the 10 topics, we provide links to well-known tutorials and hands-on materials ( Table 1 ).
We suggest a set of 10 topics to ease the start for researchers and clinicians to become engaged with basic concepts in health informatics research. We provide current review articles for more in-depth reading about the specific topic and present practical hands-on material. The presented topics likewise serve as a broad overview of the medical informatics research domain but also guide individuals and their specific interests. For example, a computer scientist familiar with CDSS development could more easily connect with important aspects, such as data privacy, FHIR, and specific EHRs that are highly relevant for daily work. In contrast, medical experts can obtain an overview of behind-the-scenes technologies, like ETL processes and underlying data quality approaches that are finally visualized as a summarizing clinical dashboard. For readers, we provided a first step toward an improved understanding of a lively and quickly expanding field, but more novel technologies and practical knowledge are ahead. Suggestions and contributions to improve the current topics can be made at GitHub, which will likewise enable content and readers to stay current [ 12 ].
Acknowledgments
This work was supported by the Federal Ministry of Health (BMG) and the German Federal Ministry of Education and Research (BMBF) within the Medical Informatics Initiative Medical Informatics in Research and Care in University Medicine (MIRACUM) Consortium (FKZ: 01ZZ180L [Dresden]; FZK: 01ZZ180A [Erlangen]; FKZ: 01ZZ1801M [Greifswald]). The article processing charge was funded by the joint publication funds of the Technische Universität (TU) Dresden, including the Carl Gustav Carus Faculty of Medicine; Saxon State and University Library (SLUB) Dresden; and the Open Access Publication Funding of the German Research Foundation (DFG).
The funding sources had no involvement in the conduct of the research and preparation of the article.
Abbreviations
Authors' Contributions: MW conceptualized the study, curated the data, and wrote the original manuscript draft. MW also defined the initial topics 1 and 2; MZ defined the initial topics 3 and 4; YP defined the initial topics 5 and 6; IR defined the initial topics 7 and 8; and NA defined the initial topics 8, 9, and 10. MS provided the resources and supervised the study. The topics were revised and extended by KF, AK, SG, DK, KLH, ICJ, CS, JS, TS, PS, and DW. MW, NA, YP, MZ, IR, and MS performed the formal analysis, and MW, NA, and MS created the visualizations. NA, YP, MZ, IR, and MS wrote, reviewed, and edited the manuscript, and all authors read and agreed to the final version of the manuscript.
Conflicts of Interest: None declared.
Qbio honours programme
- Qbio Research Proposal
- Qbio Symposiums
- Graduate School of Life Sciences
- Graduate School of Natural Sciences
- Qbio symposiums
QBio Research proposal
Instructions.
Bioinformatics Core
Grant Proposal Support
Grant Proposal Assistance and Research Collaborations
The Bioinformatics core provides the following research and grant support services to MSU faculty, staff and students:
Grant application support
Experimental design and methods consultation.
We’re happy to provide you with advice on the bioinformatic and data science aspects of your research proposal. Email [email protected] to set up a free consultation to discuss your needs and provide information about experimental design, relevant bioinformatics methods, and budget planning.
Letters of support
Email [email protected] to request a letter of support from the Bioinformatics core for your grant application.
Facilities statement
Sample language for including the Bioinformatics Core in the Facilities and Resources section of NSF and NIH grant applications is available in PDF and Word format. We can work with you to tailor a more specific statement to your needs.
Long-term research collaborations
Investigators that are interested in a Bioinformatics Core collaboration that will be part of a grant proposal or other long-term project should contact Dr. Brian O’Shea at least a few weeks before the final grant budget is due. The goal of these discussions will be to define the scope of the work that will be done, deliverables, an approximate time scale, the personnel that will be involved (which may include multiple people, depending on needs), and the estimated number of hours of effort that will be required for the Bioinformatics Core staff in order to achieve these goals. After consensus is achieved, this will be codified in a Memorandum of Understanding (MoU) between the Director, the Bioinformatics Core staff involved, and the investigator(s) in question. Upon request, it can also be documented in a letter of commitment or formal estimate of costs and services that can be included in a grant proposal.
When a project is underway, Bioinformatics consultants will keep track of the total time that they have committed to a project and the general categories of their effort (e.g., work done in pursuit of the project’s goals, meetings with other project members, training in new tools/technologies/methods). This time, along with a summary of what has been done and an estimate of progress toward the goals documented in the MoU, will be reported to the project PI and Director on a quarterly basis via email. In general, any effort that directly supports the project goals, including meetings with project personnel, will be billable to the project. A reasonable amount of time taken to learn new tools/technologies/methods/etc. in direct support of the project will also be billable, but this is expected to be a relatively small fraction of the overall project expense (no more than 25% of the total direct cost of the project). Additional time devoted to skill development by Bioinformatics core consultants will count as an in-kind contribution by the core, at no cost to the project.
We acknowledge that it is difficult to precisely estimate the amount of time that research-related efforts will require, and that grant budgets are typically fixed (i.e., cost overruns are either impractical or impossible). If, during a collaborative project, it becomes clear that the budgeted effort is insufficient to meet the agreed-upon project goals we will have a discussion with the project PI, Bioinformatics core staff, and director to determine a path forward. The Core’s primary objectives in such a discussion are to ensure that the most crucial goals of the project are met while also maintaining the ability of Bioinformatics Core personnel to complete other crucial aspects of their work. Once these discussions have been concluded, the outcome will be documented via an amended MoU.
2025 Delta Research Awards: Proposal Solicitation

IMPORTANT DATES
April 19, 2024 10:00AM PDT: RFP Informational Webinar (Optional)
May 14, 2024 5:00PM PDT: Letter of Intent due to eSeaGrant
June 14, 2024 10:00AM PDT: Proposal Preparation Webinar (Optional)
August 26, 2024 5:00PM PDT: Full proposals due to eSeaGrant
December 2024: Intent to Award issued
April 1, 2025: Expected project start date
Proposals will only be accepted from applicants whose Letters of Intent have been approved and who have received an invitation to submit a full proposal.
Table of Contents
- What's new about this Solicitation?
- Where to Find Help
- Award Information and Project Categories
- Submittal Requirements
- Eligibility Requirements
- Solicitation Focus
- Proposal Requirements
- Proposal Review Procedure
- Resources for Applicants
1. Background
The Delta Stewardship Council (Council) is pleased to announce the 2024 Delta Research Awards Proposal Solicitation. This proposal solicitation for Delta research projects (Solicitation) is funded by the Council, led by the Council’s Delta Science Program (DSP), and administered in partnership with the University of California San Diego, California Sea Grant (Sea Grant). The Solicitation will further the DSP’s legislatively mandated mission to:
… provide the best possible unbiased scientific information to inform water and environmental decision-making in the Delta … through funding research, synthesizing and communicating scientific information to policy-makers and decision-makers… -Delta Reform Act 2009, Water Code Section 85280(b)(4).
Through this Solicitation, the DSP seeks to identify and fund research that will promote an integrated understanding of the Sacramento-San Joaquin Delta and Suisun Marsh, particularly to support the science and natural resource management community’s ability to measure, anticipate, and plan for a rapidly changing climate. Proposals must advance one or more of the Science Actions in the 2022-2026 Science Action Agenda (SAA) . The SAA prioritizes science actions to fill gaps in knowledge and aligns them with management needs. For more information about the Solicitation focus and the SAA, see Section 8 .
Eligible entities that wish to submit a proposal must first submit a Letter of Intent by the deadline set forth in the Solicitation as a prerequisite to be considered for an invitation to submit a full proposal. Letters of Intent will be evaluated based on the requirements in Section 6.1 of the Solicitation and successful applicants will receive a notification to submit a proposal. All proposals will be evaluated by independent experts with the appropriate specialized knowledge, based on requirements and criteria in Sections 9 and 10 of the Solicitation. The Council will select proposals for final awards. Selected applicants will receive an “intent to award” letter and will be required to enter into a contract agreement (agreement) to be negotiated with Sea Grant. If additional funding is available from external partners, successful proposals may receive an “intent to award” letter from the Council and/or external funding partners such as the Bureau of Reclamation and State Water Contractors, as applicable. There is a total of approximately $6 million available for awards. Sea Grant will collaborate closely with the Council in administering the Solicitation as well as for external and expert review of submitted proposals, award agreements, and communication of funded work with key interested parties.
2. What's new about this solicitation?
- There are separate award categories for large projects ($200,001 to $1,500,000) and small projects ($90,000 to $200,000). The category for small projects was added following public input on the 2021 Solicitation.
- Projects must directly advance at least one science action from the 2022-2026 SAA.
- In recognition of the importance of SAA actions related to the human dimensions of the Delta, projects with a substantial social science component will be eligible for additional points during the review process ( Section 10 ). Data from the 2023 Delta Residents Survey may be relevant to researchers (Section 11.2).
- Letters of Intent will be assessed based on whether the proposed project aligns with science actions identified in the 2022-2026 Science Action Agenda, meets eligibility criteria, and falls within the geographic scope of the Delta (Section 6.1).
- Large projects are required to have one or more Letter(s) of Support from a Delta community partner, resource manager, or decision-maker (Section 9.6).
- All awards will be administered as formal agreements with Sea Grant. All collaborating entities will also be required to enter into sub-agreements with the primary applicant or may be required to enter into a separate agreement with Sea Grant.
- For optional assistance identifying tribal and/or community partners, please submit a Partnership Survey response here by May 1, 2024: https://www.surveymonkey.com/r/N7X8S9F
3. Schedule
Table 1. Schedule in Pacific Daylight Time (PDT)
Schedule is subject to change. Updates will be sent to applicants who have submitted a Letter of Intent via the eSeaGrant online portal .
4. Where to Find Help
Please see this website for the most updated copy of the Solicitation , answers to questions, and other information about the Solicitation and proposal process. For important resources and links, reference Section 11 , Resources for Applicants.
For technical assistance and questions about the Solicitation, please contact [email protected].
Communications with Council or Sea Grant staff related to the Solicitation, other than as specified and allowed in the Solicitation, may disqualify a potential proposal from being considered. To ensure that your questions will be answered in a timely manner, we recommend sending questions relating to proposal preparation and submission prior to August 19, 2024.
Two optional virtual webinars will be held to provide technical assistance and other guidance for proposals (see Section 3 , Schedule). Additional virtual webinars and/or workshops may be held on topics relevant to this Solicitation. Applicants registered on eSeaGrant will be notified of workshop details. The information will also be posted on the Council’s events calendar web page . Workshops will be recorded, and the recordings will be made available on the Solicitation website.
5. Award Information and Project Categories
There is a total of approximately $6 million available for awards. Projects must directly advance at least one science action from the 2022-2026 SAA. Availability of funding is dependent upon State and Federal budget appropriations for the specified fiscal year and is subject to change. All awards selected by the Council will be administered as formal agreements with Sea Grant. In some cases, additional awards may be selected by, and administered as formal agreements with, external partners.
Project categories (dollar amount limits include all eligible costs including indirect costs):
- Small Projects: Awards between $90,000 and $200,000
- Large Projects: Awards between $200,001 and $1,500,000
The project duration may be up to a maximum of three years (36 months).
Applicants may submit more than one Letter of Intent and proposal (subject to receiving an invitation to submit a proposal), but a maximum of one proposal per individual lead Principal Investigator (PI) can be selected for an award. However, lead PIs may be listed as co-PIs on other awarded projects if the total combined effort of awarded projects is less than or equal to 100% of their time.
Budget Contingency Clause for State-Funded Contract Agreements
If the Budget Act of the current year and/or any subsequent years covered under the ensuing agreement does not appropriate sufficient funds for the program, the agreement shall be of no further force and effect. In this event, the Council will have no liability to pay any funds whatsoever or to furnish any other considerations under the agreement and the contractor shall not be obligated to perform any provisions of the agreement.
If funding for any fiscal year is reduced or deleted by the Budget Act for purposes of this program, the Council will have the option to either: cancel the agreement with no liability occurring to the Council or offer an agreement amendment to the contractor to reflect the reduced amount. The contractor shall be reimbursed for any completed work or work in progress at the time of termination of an executed agreement if approved by the Council.
Recognition of Funding Source
Successful applicants must acknowledge funding from the Delta Stewardship Council and its Delta Science Program, and any partner organizations providing project funds, as specified in the agreement language, for any publication (including online webpages) of any material based on or developed under a project funded through this Solicitation. Support must also be orally acknowledged during all news media interviews, including radio, television, and news magazines.
6. Submittal Requirements
Letter of intent (loi).
Letters of Intent (LOI) are required and must be submitted by the deadline in Section 3 (Schedule) using eSeaGrant: http://eseagrant2.ucsd.edu/. If you have never used California Sea Grant's eSeaGrant portal before, you will need to register for an account. You can change the randomly-generated password once you log in successfully into the website. Contact [email protected] with any access issues related to eSeaGrant. NOTE: We advise not to wait until the last minute to submit your LOI; when eSeaGrant experiences high user traffic, you may experience page loading delays. It is the applicant’s responsibility to get all required materials submitted before the deadline, and the submission deadline will not be extended.
All interested applicants must submit a Letter of Intent (LOI), which contains a brief description of their project, using eSeaGrant by the deadline specified in the Solicitation (see Section 3, Schedule). For projects with multiple collaborating entities requesting funds, one lead PI should submit a single LOI on behalf of all collaborating entities. LOIs will be used to screen for eligibility and relevance to the Science Action Agenda, to enable the timely selection of reviewers, and to help avoid potential conflicts of interest in the review process. Interested applicants may submit more than one LOI, but an individual may only be the Primary Investigator for a single submitted project.
LOIs will be screened based on the requirements below. An invitation to submit a proposal will be issued to each applicant whose LOI passes the screening process. LOIs received after the deadline will not be considered.
If there are any proposed changes to the scope of the successful LOIs, applicants must notify California Sea Grant via [email protected] as soon as possible and no later than July 15th, 2024. Proposed changes may only include changes in the lead PI/institution or contact information, co-PI(s), budget, award type sought (large/small), geographic scope, and the approach including which SAAs will be addressed. Applicants will be notified by email no later than July 23rd, 2024 regarding whether the changes to their LOI are accepted, including an invitation to submit a proposal (if applicable) with the accepted revision(s).
LOIs will be assigned a pass/fail score based on their relevance to the science actions identified in the 2022-2026 Science Action Agenda ( Section 8 , Solicitation Focus), eligibility ( Section 7 , Eligibility Requirements), and whether they fall within the geographic scope of the Delta. Projects are not required to be physically located within the Delta; however, project activities must provide a demonstrable link(s) to the Delta. A link to the Delta could include hydrologic connection, tribal ancestral/spiritual connection, social/cultural connection, etc. The ‘Delta’ means the Sacramento-San Joaquin Delta as defined in Water Code Section 12220 and the Suisun Marsh as defined in Public Resources Code Section 29101 (Water Code Section 85058).
Applicants will be notified electronically in writing if their LOIs were or were not successful. Applicants with successful LOIs will receive an electronic invitation to submit a full proposal. Applicants that did not receive an invitation to submit will not be considered.
The page limit for the LOI is two (2) pages, Arial font size 12, single spacing, and standard margins, including header, footer, labeling, and address information. If the LOI exceeds two pages, only information in the first two pages will be considered.
LOIs must include the following information submitted through forms in eSeaGrant:
- Name of lead PI, affiliation, and contact information (name of lead PI must not change from LOI to proposal submission).
- Name of Co-PI(s) with affiliation(s), if applicable.
- Title of project.
- Indication of award type sought (Large Project or Small Project, see Section 5 , Award Information and Project Categories) and which SAA Science Action(s) will be addressed.
- Geographic scope of the project.
- Brief discussion of the topic and approach, including how the specified science action(s) will be addressed.
- Approximate total budget and a list of all the collaborating entities who will receive funds as part of the award.
Project proposal
Proposals will only be accepted from applicants whose Letters of Intent have been approved and who have received an invitation to submit a full proposal. Applicants who do not receive an invitation to submit a proposal will not be considered.
All proposals must present clear hypotheses or cogent research questions that can be addressed using a scientifically-sound research design. Research may invoke disciplines within, for example, the biophysical sciences, social sciences, integrated social-ecological disciplines, traditional knowledge, and/or local place-based knowledge.
Proposals are encouraged to:
- Include substantial roles for undergraduate, graduate, and/or postdoctoral students, particularly those from underrepresented groups and a diversity of lived experiences;
- Have a plan for meaningful, early, and sustained engagement with community members or community organizations;
- Be based on or thoughtfully and respectfully incorporate tribal, traditional, and/or local knowledges, as applicable.
- Proposals must meet all the requirements in Section 9 (Proposal Requirements) and must be submitted by the deadline in Section 3 (Schedule) using eSeaGrant: http://eseagrant2.ucsd.edu/. If you have never used California Sea Grant's eSeaGrant portal before, you will need to register for an account. You can change the randomly-generated password once you log in successfully into the website. Contact [email protected] with any access issues related to eSeaGrant. NOTE: We advise not to wait until the last minute to submit your proposal; when eSeaGrant experiences high user traffic, you may experience page loading delays. It is the applicant’s responsibility to get all required materials submitted before the deadline, and the submission deadline will not be extended.
7. Eligibility Requirements
Eligible entities.
All entities will be required to fulfill the award conditions of the University Terms & Conditions (UTC-220) and all pass-through terms and conditions from the Council unless otherwise agreed upon by the parties.
Eligible entities for agreements are entities that are in good standing and eligible to do business in California, including but not limited to:
- A California Native American Tribe;
- A California State agency, State college, or State university, including an auxiliary organization of the California State University (CSU);
- A State agency, State college, or State university from another state;
- A local governmental entity, including those created as a Joint Powers Authority and local government entities from other states;
- California community colleges including an auxiliary organization or foundation organized to support the Board of Governors of the California Community Colleges;
- The Federal government including National Laboratories;
- An auxiliary organization of the Student Aid Commission established under Education Code;
- A corporation (both domestic and foreign), partnership, limited partnership, or limited liability company, or other such similar organization that meets the requirements for doing business in California, including tax-exempt organizations such as 501(c)(3) non-profit organizations;
- A private independent business, including sole proprietors;
- A domestic or foreign private college, university, or educational or research entity.
For proposals involving multiple entities, a single entity must be identified as the primary lead entity, and a single proposal describing the entire project must be submitted by that entity. The budgets of those participating entities must be clearly identified in the comprehensive project budget submitted by the lead entity and not exceed the total project budget.
Eligible activities include, but are not limited to:
- Research and data collection, analysis, synthesis, management, and delivery;
- Development of resource management tools and technologies;
- Development of conceptual or quantitative models;
- Production of peer-reviewed journal articles, conference presentations, and communications for the scientific/management community;
- Science communication for broader audiences and/or community engagement;
- Project management and coordination of a multidisciplinary team;
- Institutional Review Board (IRB) review;
- Document/report preparation.
Ineligible Activities
Funds shall not be expended to pay:
- the design, construction, operation, mitigation, or maintenance of restoration projects or any Delta Plan covered actions , or
- implementation activities (e.g., construction or improvement of a capital asset), or
- land acquisition or easement purchase, or
- information technology (IT) services (e.g., hardware, software, web services) as defined: https://www.dgs.ca.gov/PD/Resources/SCM/TOC/10/10-2
See Proposal Requirements Section for Ineligible Costs.
8. Solicitation Focus
Proposals must directly address one or more of the 25 priority science actions described in the 2022-2026 SAA and must either be physically located in the Delta or provide a demonstrable link to the Delta. The ‘Delta’ means the Sacramento-San Joaquin Delta as defined in Water Code Section 12220 and the Suisun Marsh as defined in Public Resources Code Section 29101 (Water Code Section 85058). A link to the Delta could include hydrologic connection, tribal ancestral/spiritual connection, social/cultural connection, etc.
In the Solicitation Notice , the section, “Solicitation Focus,” provides a high-level summary of the SAA, listing actions under thematic management needs. The management needs and the science actions are of equal priority and not listed in order of importance and are cross-cutting and integrative and unlikely to be addressed by only one project. More points will be awarded to projects that address multiple components of a science action or multiple science actions, where appropriate. For more information about the 25 priority science actions that are the focus of this Solicitation, please review the full SAA document.
Science Actions:
- Establish publicly accessible repositories, interactive platforms, and protocols for sharing information, products, and tools associated with monitoring and modeling efforts, in support of forecast and scenario development, timely decision-making, and collaborative efforts.
- Evaluate the individual and institutional factors that enable or present barriers to coordination, learning, trusting, and using scientific information to inform decision-making and resource sharing within and among organizations.
- Identify and implement large-scale experiments that can address uncertainties in the outcomes of management actions for water supply, ecosystem function, and socioeconomic conditions in the Delta.
Science Actions:
- Evaluate and update monitoring programs to ensure their ability to track and inform the management of climate change impacts, emerging stressors, and changes in species distributions.
- Develop a framework for monitoring, modeling, and information dissemination in support of operational forecasting and near real-time visualization of the extent, toxicity, and health impacts of Harmful Algal Blooms (HABs).
- Enhance flood risk models through a co-production process with Delta communities to quantify and consider tradeoffs among flood risk management, water supply and water quality management, habitat restoration, and climate adaptation.
- Iteratively develop, update, and make widely available forecasts of climatological, hydrological, social-ecological, and water quality conditions at various spatial and temporal scales that consider climate change scenarios.
- Conduct studies to inform restoration and approaches to protecting human communities that are resilient to interannual hydrologic variation and climate change impacts.
- Develop integrated frameworks, data visualization tools, and models of the Delta social-ecological system that evaluate the distribution of environmental benefits and burdens of management actions alongside anticipated climate change impacts.
- Identify how ecosystem restoration projects, in comparison to existing water management strategies, benefit and burden human communities, with an emphasis on environmental justice.
- Test and monitor the ability of tidal, nontidal, and managed wetlands and inundated floodplains to achieve multiple benefits over a range of spatial scales, including potential management costs, tradeoffs, and unintended consequences.
- Synthesize existing knowledge and conduct applied, interdisciplinary research to evaluate the costs and benefits of different strategies for minimizing the introduction and spread of invasive species, and to inform early detection and rapid response strategies.
- Use multi-method approaches (e.g., surveys, interviews, oral histories, and/or observations) to develop an understanding of how human communities’ values, and uses of cultural, recreational, agricultural, and natural resources vary across geography, demographics, and time.
- Synthesize existing data and collaboratively develop additional long-term data collection and monitoring strategies to address knowledge gaps on human communities within the Delta and those reliant on the Delta, with the goal of tracking and modeling metrics of resilience, equity, and well-being over time.
- Measure and evaluate the effects of using co-production or community science approaches (in management and planning processes) on communities' perceptions of governance and on institutional outcomes, such as implementation or innovation.
- Identify and test innovative methods for effective control or management of invasive aquatic vegetation in tidal portions of the Delta under current and projected climate conditions.
- Identify thresholds in the survival and health of managed fish and wildlife species with respect to environmental variables (e.g., flow, temperature, dissolved oxygen) and location-specific survival probabilities to develop strategies that will support species recovery.
- Determine how environmental drivers (e.g., nutrients, temperatures, water residence time) interact to cause HABs in the Delta, identify impacts on human and ecosystem health and well-being, and test possible mitigation strategies.
- Integrate and expand on existing models of hydrodynamics, nutrients, and other food web drivers to allow for the forecasting of the effects of interacting stressors on primary production and listed species.
- Quantify spatial and temporal patterns and trends of chemical contaminants and evaluate ecosystem effects through monitoring, modeling, and laboratory studies.
- Evaluate how climate change, sea level rise, and more frequent extremes will impact habitats, water supply, water quality, sediment supply, long-term species persistence, primary productivity, and food webs.
- Evaluate individual and cumulative impacts and tradeoffs of drought management actions on ecological and human communities over multiple timescales.
- Evaluate the possible multi-benefits of management actions that promote groundwater recharge for ecological functions and water resilience under climate change (e.g., multiple dry year scenarios).
- Identify how human communities connected to the Delta watershed are adapting to climate change, what opportunities and tradeoffs exist for climate adaptation approaches (i.e., agricultural practices, carbon sequestration, nature-based solutions/green infrastructure), and how behaviors vary with adaptive capacity.
- Predict and test how water allocation and supply decisions, and ecological flow scenarios should change under projected climate change to maintain habitat conditions, access of target species to critical habitat, and interactions among native and invasive species.
9. Proposal Requirements
Eligible entities that wish to submit a proposal must first submit a LOI by the deadline set forth in the Solicitation as a prerequisite to be considered for an invitation to submit a full proposal (See Section 3 , Schedule).
Applicants with successful Letters of Intent will receive an electronic invitation to submit a full proposal. The invitation to submit must be included with the proposal submittal.
Listed below are the requirements for a complete proposal package; for full details on each component, please refer to the Proposal Solicitation Notice . For lead PIs affiliated with academic institutions, final proposals must be submitted by the institution’s sponsored research office. For deadlines, see Section 3 (Schedule). For instructions on how to submit a proposal via eSeaGrant, see Section 6.2 (Project Proposal). For award information, see Section 5 (Award Information and Project Categories).
10. Proposal Review Procedure
Each proposal submitted by the deadline specified in Section 3 will undergo several steps in the review and selection process:
- Proposals will be screened in an administrative review by Sea Grant;
- Proposals that pass the administrative review will be advanced to a technical review by subject matter experts ( individual expert technical reviews );
- Individual expert technical reviews will be considered during one or more technical evaluation panel(s) during which the proposals will be reviewed, discussed and ranked;
- The Council, in consultation with the Delta Lead Scientist, will make funding decisions based on consideration of the technical reviews, rankings, and factors described in Section 10.4, Funding Decisions.
Further details on each of these steps are below.
Administrative Review
Administrative review determines if the proposal meets the following criteria:
- The applicant and project are eligible. See Section 7 , Eligibility Requirements, for eligibility requirements.
- The proposal is complete. The proposal has all required sections: see Section 9 , Proposal Requirements.
Proposals that do not meet both criteria may not be considered eligible under this Solicitation.
Individual Expert Technical Reviews
All proposals that advance past administrative review will go through independent technical review by at least two external experts. Technical reviewers will be professionals in fields relevant to the proposed project and screened to avoid any potential conflicts of interest. Technical reviewers will evaluate each proposal in accordance with the Technical Review Criteria (Table 2) and may submit narrative comments that support their scores.
Table 2. Technical Review Criteria
The following is a list of questions that will be provided as guidance for proposal reviewers:
- Will the work address key scientific uncertainties and fill important information gaps? The proposed research does not have to be hypothesis-driven but must, at a minimum, include a clear statement of research questions.
- Is the underlying scientific basis or underlying knowledge base for the proposed work clearly explained, the need for the project justified, and is it based on the best possible information, such as current scientific literature, Tribal expertise, traditional knowledge, and local knowledge?
- Are the methods, including data analysis and reporting, clearly linked to and appropriate for the objectives and research questions?
- How is the project responsive to the 2022-2026 SAA? Which science action(s) will be addressed? Does the project address more than one science action? How comprehensively does the project address the science action(s)?
- Large Projects Only: Does the letter of support demonstrate an effective connection with management needs and meaningful engagement with practitioners, Delta communities, and/or resource managers?
- Is the proposed work significant on the landscape and regional scale?
- Will the information produced contribute to effective adaptive management or co-production (i.e., participatory knowledge development) of science for the Delta?
- If applicable: Will the project leverage existing datasets or tools?
- Is there evidence that the project team has made good faith efforts to engage with community groups or Tribes?
- How well does the proposed project incorporate realistic and ample opportunities for community partnership, participation, and/or input?
- How will feedback from engagement be incorporated into or influence the proposed work?
- Will there be any co-production of knowledge or participatory research with tribal experts or community groups?
- Will the research process and/or products have the potential for a meaningful positive impact on underrepresented groups or to promote EJ?
- Will the process and /or products promote principles of justice, equity, diversity, and inclusion?
- Does the Engagement and Communication Plan explain how the information will be made directly available to the entities that will most benefit from it, including scientists, managers, and the public?
- Does the proposed work include training and mentoring for students (K-12, undergraduate, graduate), post-doctoral scholars, and/or educators (e.g., curriculum development), particularly those from underrepresented groups and with a diversity of lived experiences?
- Is there a plan for policy engagement, such as presentations to decision-makers?
- Will the proposed work include partnerships among academic, industry, and/or non-governmental organizations?
- Does the proposed project employ methods, theories, or data from any of the social science disciplines, including but not limited to political science, sociology, economics, anthropology, geography, or psychology?
- Does the project meaningfully integrate information on social and natural dimensions of the Delta?
- Is there an adequate description of how each element of the project will be implemented (e.g., methods, materials, equipment, responsible parties)?
- Does the schedule demonstrate a logical sequence and timing of project tasks? Is it feasible to complete the proposed work within the proposed time frame? Are potential pitfalls and contingencies described in sufficient detail?
- Are the necessary facilities, equipment, and administrative capacity available to successfully perform and manage the proposed tasks?
- Is there justification for all costs in the budget?
- Are all costs well justified and realistic for the work being proposed?
- Does the project team have adequate expertise to complete the proposed work?
- What is the project team’s record of publication, productivity, management, engagement, training, and outreach?
- Does the DMP address all sections described in the Solicitation, including best practices for open science?
Technical Evaluation Panel(s)
The Review Panel(s) will consider the individual reviews by technical experts and rank projects according to the review criteria listed in the Individual Expert Technical Reviews Section. Members of the review panel(s) will be professionals in fields relevant to the proposed projects and screened to avoid any potential conflicts of interest.
Funding Decisions
The Council will select proposals for awards in consultation with the Delta Lead Scientist (or if the Delta Lead Scientist Position is vacant, the Deputy Executive Officer for Science or the Deputy Executive Officer for Science’s designee). Funding decisions will be made with consideration of the following:
- Review Panel feedback and rankings
- Distribution of projects across SAA science actions
- Budget requests relative to available funds
- Management relevance to the Delta
- Distribution of applicants’ institutions and career stages
Any funding partners will select proposals in coordination with the Council and issue intent to award letters separately.
The intent to award does not guarantee an ensuing agreement. For proposals recommended for funding, intent to award letters will be distributed to the primary applicant and will include any requested changes to the proposal and/or budget in response to proposal review feedback. The Council reserves the right to revise funding decisions. To proceed to an executed agreement, successful applicants must provide any revisions and additional documentation as requested by Sea Grant in a timely manner.
11. Resources for Applicants
Please see the funding solicitation PDF for a full list of resources relating to:
- Science Action Agenda
- Delta Residents Survey Data
- Environmental Justice
- Community Engagement
- Data Management
- More About the Delta Stewardship Council
- State and Regional Resources
- Definitions
Appendix A: Award Reporting Template
Appendix B: Budget Template
Appendix C: Engagement and Communication Template
Application Resources
Subscribe to california sea grant emails, subscribe to california sea grant emails.
- Funding, fellowship, and job announcements
- Russian River Salmon & Steelhead Program
- Delta Stewardship Council Funding/Fellowships
View previous campaigns.
Opinion | Biden administration proposal threatens…
Share this:.
- Click to share on Facebook (Opens in new window)
- Click to share on Twitter (Opens in new window)
- Click to share on Reddit (Opens in new window)
- Click to print (Opens in new window)
- Opinion Columns
- Guest Commentary
- Letters to the Editor
- Editorial Board
- Endorsements
Opinion | Biden administration proposal threatens innovative research at universities across the country

UCLA just purchased a 700,000-square-foot property in Westwood that it’s planning to remodel into a state-of-the-art research park for quantum science, immunology, immunotherapy, and other high-tech fields. UCLA has billed the park as the “future home of discoveries that will change the world.”
Despite such visionary local leadership, however, policymakers in Washington are poised to scuttle innovation at universities across the country. The Biden administration plans to reinterpret a decades-old law, the Bayh-Dole Act, that is at the heart of university-based research and development.
The proposal would affect patents on any invention arising from federally funded research. It asserts the federal government’s supposed authority to “march in” and effectively seize patents when officials think a product’s price is too high.
In essence, the federal government wants to control the price of university-based innovations. Doing so would blow up the “technology transfer” system that turns breakthrough discoveries into real solutions. Products on the chopping block include life-saving therapies and quantum computers.
This would set us back to before 1980, when the government maintained control over all patents associated with federal funding. Because Washington had neither the capacity nor incentive to commercialize these inventions, and universities cannot make and sell products on their own, publicly funded breakthroughs rarely yielded tangible benefits.
Bayh-Dole solved this problem by allowing universities and other federally funded research institutions to retain patent rights for their discoveries. That enabled them to partner with private businesses that bring their inventions to market. In turn, universities collect royalties that support more students and more research, creating a continuous cycle of innovation.
Bayh-Dole unlocked the vast innovation potential of America’s universities. Before Bayh-Dole, federally funded research had produced roughly 30,000 patents, but the government had licensed fewer than 1,500 for commercialization. In comparison, 2022 alone saw nearly 17,000 patent applications filed for federally funded discoveries and almost 10,000 licenses executed. The Act supports millions of jobs, has helped launch over 17,000 start-ups, and has contributed around $2 trillion to U.S. output.
UCLA’s new research park helps illustrate Bayh-Dole’s influence. Google, which supported UCLA’s acquisition of the site, was founded to commercialize a patented search engine algorithm from Stanford University. Meanwhile, it was a revolutionary drug developed by UCLA faculty that sparked the launch of the field of cancer immunotherapy, a primary focus of the new park.
Private sector partners are critical for bringing such university innovations to market, and they rely on patents to justify their investment. If the government casts doubt on the reliability of these patents, firms will hesitate to license and develop early-stage research. Unfortunately, the new patent seizure plan will do just that.
The administration maintains it will only exercise this newfound authority when prices are “unreasonable,” whatever that means. But if the government can decide the level of profitability, especially based on such arbitrary, unpredictable standards, the private sector will avoid all promising inventions generated from federal funds. In the end, they will not reach the public.
Not only is the proposal bad policy, it is also illegal. The Bayh-Dole Act does not give the administration price-control authority. In fact, the law’s bipartisan architects, Senators Birch Bayh and Bob Dole, explicitly cautioned against it. And every single presidential administration, from both parties, has consistently declined to use the law to regulate prices.
UCLA envisions the new research park as “bring[ing] scholars from different higher education institutions, corporate partners, government agencies and startups together to…achieve breakthroughs that will serve our global society.” This type of cooperation has become the norm under Bayh-Dole. It will end abruptly if the Biden administration rewrites the rules of the game.
Fortunately, there are better approaches to improving access to drugs and other technologies. UCLA, for example, recently partnered with the UN’s Medicines Patent Pool and the student-led UAEM (Universities Allied for Essential Medicines) to require that licenses include an Affordable Access Plan for low- and middle-income countries. Leaving the crafting of such plans to private-public partnerships makes more sense than Washington big-footing it.
UCLA is investing $500 million in developing the new research park. The private sector will add much more. But for these investments to ultimately benefit the public, the Biden administration must lay off Bayh-Dole.
Amir Naiberg serves as associate vice chancellor and president & CEO of UCLA Technology Development Corporation. Andrei Iancu served as the undersecretary of Commerce for intellectual property and director of the U.S. Patent and Trademark Office from 2018 to 2021 and serves as board co-chair of the Council for Innovation Promotion.
- Newsroom Guidelines
- Report an Error
More in Opinion

Opinion | Rent control is ‘a ludicrous idea,’ and so is blocking housing

Opinion | Norma Torres vs. a QAnoner, part three

Opinion | Proposition 28 funding, intended to supplement arts funding in schools, is being misused

Opinion | In search of the ‘Tomato King’: Finding a Mexican migrant politician, rooted in California soil

IMAGES
VIDEO
COMMENTS
If more information is needed, please contact: Drena Dobbs Dept. of Zoology and Genetics 2114 Molecular Biology Building Iowa State University Ames, IA 50011. (515)294-1112 [email protected]. PROPOSAL FOR AN INTERDEPARTMENTAL MAJOR. IN BIOINFORMATICS AND COMPUTATIONAL BIOLOGY.
This platform, originally conceived as a research tool, integrates a community-driven teaching framework with a wide collection of training materials covering diverse bioinformatics domains [63,64]. Some practical recommendations for using Galaxy as an e-learning platform have been compiled in Serrano-Solano and colleagues [ 11 ].
An important component of providing effective bioinformatics support is conducting research that is reproducible and reusable. When conducting data analysis, it is crucial to employ appropriate bioinformatics methods (tools and resources) and statistical models that deliver reliable inferences from the data. As is the nature of the science ...
It is noted by the authors of this piece, that the advent of blended/ online learning approaches holds the potential to resolve many of the points raised here. Some general guidelines for virtual ...
A recent study has found that the interest of researchers in these topics plateaued over after the early 2000s [1]. Besides the above mentioned hot topics, the following topics are considered demanding in bioinformatics. Cloud computing, big data, Hadoop. Machine learning. Artificial intelligence.
These articles reflect current trend and development in bioinformatics research. The supplement to BMC Bioinformatics was proposed to launch during the BIOCOMP'19—The 2019 International Conference on Bioinformatics and Computational Biology held from July 29 to August 01, 2019 in Las Vegas, Nevada. In this congress, a variety of research ...
PhD studentship in Population Health Sciences: Developing machine learning models for integration of molecular and clinical data. Newcastle University Population Health Sciences Institute. Award summary . 100% tuition fees (paid at home rate) and a minimum annual stipend of £19,237 (2024/2025) with support for research costs.
The interdisciplinary nature of bioinformatics makes it an ideal framework to develop activities enabling enquiry-based learning. We describe here the development and implementation of a pilot project to use bioinformatics-based research activities in high schools, called "Bioinformatics@school." It includes web-based research projects that students can pursue alone or under teacher ...
Bioinformatics and Computational Biology Core (15 Credit Hours) Bioinformatics: BINF644 Bioinformatics (3) Systems Biology [Select One] ... Following completion of the research outlined in the proposal, the MS degree candidates will prepare a written thesis according to the guidelines set forth by the Graduate College. A thesis defense ...
PhD proposal in Bioinformatics AphidRNA: Learning and modeling a RNA interaction network associated to the pea aphid reproduction modes Research teams: SYMBIOSE (Bioinformatique) Centre INRIA Rennes + Biologie et Génétique des Populations d'Insectes INRA BIO3P.
The Bioinformatics Programmatic Area supports the design of novel and innovative bioinformatics approaches that have the potential to become part of the cyberinfrastructure that will advance or transform biological understanding and that has the potential to be broadly applicable in biology. Proposed projects may be focused on any biological ...
Briefings in Bioinformatics welcomes proposals for special themed issues! Special issues normally comprise eight to ten articles of up to 5,000 words each. Suggestions or proposals for special themed issues should be addressed to the editorial office ( [email protected]) in the first instance. Briefings in Bioinformatics welcomes ...
This research will endeavor to incorporate automation and bioinformatics with AI(Artificial Intelligence), which will substitute the unnecessary interactions with medical practitioners and usher the medical profession into the new age of automation through AI bioinformatics and health informatics amalgamation using Artificial Intelligence.
Purpose of Review This review article provides an overview of the bioinformatics frameworks developed in recent years that will facilitate analysis and promote the application of stem cells to medical research. Recent Findings High-throughput profiling techniques at the transcript, epigenetic, and proteomic level have uncovered unique molecular signatures of stem cells that underlie their ...
Abstract. The currently emerging pathogen SARS-CoV-2 has produced the global pandemic crisis by causing COVID-19. The unique and novel genetic makeup of SARS-CoV-2 has created hurdles in biological research, due to which the potential drug/vaccine candidates have not yet been discovered by the scientific community.
We are happy to provide you with assistance on the bioinformatic and data science aspects of your proposal. We are happy to arrange a free consultation to discuss the needs of your proposed project and to provide support, including: experimental design. relevant bioinformatics methods. budget planning. facilities and resources statements.
Research Proposal - Bioinformatics approach to evaluation of Transcription factor genes and diseases (Cancer) - Free download as Word Doc (.doc), PDF File (.pdf), Text File (.txt) or read online for free. The purpose of the proposed research is the development of a computational approach to quantitatively evaluate associations between transcription factor encoding genes and human diseases ...
Looking for examples of NIH proposals in the field of Bioinformatics. I am a senior scientist in the field of bioinformatics but I never wrote my own research proposal, and having an award would definitely help me to transition to a professor position. I have some ideas in mind but it is difficult for me to put these ideas in a proposal format ...
Topic Selection. The initial topics were defined based on current developments in the health informatics field and an increasing number of published manuscripts between 2000 and 2021 (based on title-abstract-keyword screening in Scopus using the keywords "Health" AND "Informatics" AND "domain") in the respective subdomains (Figure 1 A).). After a first definition of the specific ...
QBio Research proposal. ... (such as bioinformatics, mathematical modelling and/or computer simulations) over the whole spectrum of the Life Sciences. The proposal should be prepared using the form provided, and needs to be submitted before 3 July 2017 (by email to Can Kesmir). The main section containing the research proposal (section 6) has a ...
Postgraduate Course: Research Proposal (Bioinformatics) (PGBI11114) After two introductory tutorials, the details of the project will be largely developed by the student in consultation with their project supervisor, who will provide suggestions and background reading. The aim of this course is to develop generic research skills that can be ...
Apache/2.4.38 (Debian) Server at web.media.mit.edu Port 443
Experimental design and methods consultation. We're happy to provide you with advice on the bioinformatic and data science aspects of your research proposal. Email [email protected] to set up a free consultation to discuss your needs and provide information about experimental design, relevant bioinformatics methods, and budget planning.
1. Background. The Delta Stewardship Council (Council) is pleased to announce the 2024 Delta Research Awards Proposal Solicitation. This proposal solicitation for Delta research projects (Solicitation) is funded by the Council, led by the Council's Delta Science Program (DSP), and administered in partnership with the University of California San Diego, California Sea Grant (Sea Grant).
(1) General Assembly resolution A/73/L.40/Rev.1 adopted on 13 December 2018 decided that São Tomé and Príncipe and Solomon Islands will graduate six years after the adoption of the resolution, i.e., on 13 December 2024.
The Biden administration plans to reinterpret a decades-old law, the Bayh-Dole Act, that is at the heart of university-based research and development. The proposal would affect patents on any ...
Webinar. NIJ will host a webinar on April 17, 2024, from 1-2pm ET discussion this solicitation. Register for the webinar.. With this solicitation, NIJ seeks research partnership proposals that meet the needs and missions of local justice and service provider entities — including police, corrections, courts, victim services, forensic science service providers, and community safety and adult ...