The role of imaging in the diagnosis of lung cancer in primary care
--> Bradley, Stephen (2022) The role of imaging in the diagnosis of lung cancer in primary care. PhD thesis, University of Leeds.
Background Lung cancer is the leading cause of cancer death worldwide. The UK relies more heavily upon chest x-ray than many other high income countries. Little is known about the performance of the test, the risk of cancer following negative test, consequences of ‘false negative’ results and the factors that affect how frequently chest x-ray is used. Aims 1. Determine sensitivity and specificity of chest x-ray. 2. Determine if there are differences in outcomes between patients with ‘true positive’ versus ‘false negative’ chest x-rays. 3. Determine the risk of lung cancer following a negative chest x-ray with respect to symptoms. 4. Quantify the volume of chest x-rays undertaken by English general practices and understand the extent to which variations in chest x-ray frequency are due to differences in patient populations and the practices themselves. Methods 1. Systematic review on sensitivity of chest x-ray 2. Observational study to determine sensitivity and compare stage and survival between those with ‘true positive’ versus ‘false negative’ results. 3. Cohort study to determine chest x-ray specificity and lung cancer risk following negative chest x-ray. 4. Retrospective study to quantify general practices’ chest x-rays with respect to characteristics of their patient populations and the practices. Results 1. Sensitivity was 77-80% (systematic review) and 82% (observational study). Specificity was 90%. 2. ‘False negative’ chest x-rays were not associated with adverse outcomes, although given the retrospective methodology this cannot be excluded. 3. Lung cancer risk following negative chest x-ray was <1% for all symptoms except haemoptysis (3%). 4. There was substantial variation in chest x-ray utilisation (median 34/1000 patients, IQR 26-43), with 18% of variance accounted for by recorded characteristics. Conclusions Chest x-ray does not identify ~20% of lung cancers but it continues to have a useful role. The substantial variation in rates of investigation suggest that it may be underutilised in many practices.

--> Final eThesis - complete (pdf) -->
Filename: Thesis 04 05 22unmarked.pdf

Embargo Date:
You do not need to contact us to get a copy of this thesis. Please use the 'Download' link(s) above to get a copy. You can contact us about this thesis . If you need to make a general enquiry, please see the Contact us page.

ThinkIR: The University of Louisville's Institutional Repository
Home > ETD > 3663
Electronic Theses and Dissertations
Machine learning approaches for lung cancer diagnosis..
Ahmed Mahmoud Ahmed Shaffie , University of Louisville Follow
Date on Master's Thesis/Doctoral Dissertation
Document type.
Doctoral Dissertation
Degree Name
Computer Engineering and Computer Science
Degree Program
Computer Science and Engineering, PhD
Committee Chair
Elmaghraby, Adel
Committee Co-Chair (if applicable)
El-Baz, Ayman
Committee Member
Imam, Ibrahim
Dunlap, Neal
Author's Keywords
CT; lung cancer; medical imaging; machine learning
The enormity of changes and development in the field of medical imaging technology is hard to fathom, as it does not just represent the technique and process of constructing visual representations of the body from inside for medical analysis and to reveal the internal structure of different organs under the skin, but also it provides a noninvasive way for diagnosis of various disease and suggest an efficient ways to treat them. While data surrounding all of our lives are stored and collected to be ready for analysis by data scientists, medical images are considered a rich source that could provide us with a huge amount of data, that could not be read easily by physicians and radiologists, with valuable information that could be used in smart ways to discover new knowledge from these vast quantities of data. Therefore, the design of computer-aided diagnostic (CAD) system, that can be approved for use in clinical practice that aid radiologists in diagnosis and detecting potential abnormalities, is of a great importance. This dissertation deals with the development of a CAD system for lung cancer diagnosis, which is the second most common cancer in men after prostate cancer and in women after breast cancer. Moreover, lung cancer is considered the leading cause of cancer death among both genders in USA. Recently, the number of lung cancer patients has increased dramatically worldwide and its early detection doubles a patient’s chance of survival. Histological examination through biopsies is considered the gold standard for final diagnosis of pulmonary nodules. Even though resection of pulmonary nodules is the ideal and most reliable way for diagnosis, there is still a lot of different methods often used just to eliminate the risks associated with the surgical procedure. Lung nodules are approximately spherical regions of primarily high density tissue that are visible in computed tomography (CT) images of the lung. A pulmonary nodule is the first indication to start diagnosing lung cancer. Lung nodules can be benign (normal subjects) or malignant (cancerous subjects). Large (generally defined as greater than 2 cm in diameter) malignant nodules can be easily detected with traditional CT scanning techniques. However, the diagnostic options for small indeterminate nodules are limited due to problems associated with accessing small tumors. Therefore, additional diagnostic and imaging techniques which depends on the nodules’ shape and appearance are needed. The ultimate goal of this dissertation is to develop a fast noninvasive diagnostic system that can enhance the accuracy measures of early lung cancer diagnosis based on the well-known hypotheses that malignant nodules have different shape and appearance than benign nodules, because of the high growth rate of the malignant nodules. The proposed methodologies introduces new shape and appearance features which can distinguish between benign and malignant nodules. To achieve this goal a CAD system is implemented and validated using different datasets. This CAD system uses two different types of features integrated together to be able to give a full description to the pulmonary nodule. These two types are appearance features and shape features. For the appearance features different texture appearance descriptors are developed, namely the 3D histogram of oriented gradient, 3D spherical sector isosurface histogram of oriented gradient, 3D adjusted local binary pattern, 3D resolved ambiguity local binary pattern, multi-view analytical local binary pattern, and Markov Gibbs random field. Each one of these descriptors gives a good description for the nodule texture and the level of its signal homogeneity which is a distinguishable feature between benign and malignant nodules. For the shape features multi-view peripheral sum curvature scale space, spherical harmonics expansions, and different group of fundamental geometric features are utilized to describe the nodule shape complexity. Finally, the fusion of different combinations of these features, which is based on two stages is introduced. The first stage generates a primary estimation for every descriptor. Followed by the second stage that consists of an autoencoder with a single layer augmented with a softmax classifier to provide us with the ultimate classification of the nodule. These different combinations of descriptors are combined into different frameworks that are evaluated using different datasets. The first dataset is the Lung Image Database Consortium which is a benchmark publicly available dataset for lung nodule detection and diagnosis. The second dataset is our local acquired computed tomography imaging data that has been collected from the University of Louisville hospital and the research protocol was approved by the Institutional Review Board at the University of Louisville (IRB number 10.0642). These frameworks accuracy was about 94%, which make the proposed frameworks demonstrate promise to be valuable tool for the detection of lung cancer.
Recommended Citation
Shaffie, Ahmed Mahmoud Ahmed, "Machine learning approaches for lung cancer diagnosis." (2021). Electronic Theses and Dissertations. Paper 3663. https://doi.org/10.18297/etd/3663
Since October 29, 2021
Included in
Biomedical Engineering and Bioengineering Commons , Computer Engineering Commons
Advanced Search
- Notify me via email or RSS
- Collections
- Disciplines
Author Corner
- Collection Policy
- License Agreements
- ThinkIR Electronic Resource Guide
- Submit Research
Related Links
- Guidelines for the Preparation and Processing of Theses and Dissertations (School of Interdisciplinary & Graduate Studies) ( PDF )
- University of Louisville Libraries Research Assistance and Instruction
- Nonexclusive License to Electronically Disseminate UofL ETD ( PDF )
- Data Management Guides for Theses and Dissertations
Home | About | FAQ | My Account | Accessibility Statement
Privacy Copyright
Dissertation Library
Our last decade has included the following phd dissertations, some of the exciting academic accomplishments from students and mentors of the pulmonary center:.
Claire Burgess Generation of Human Alveolar Epithelial Type I Cells From Pluripotent Stem Cells Mentor: Darrell Kotton Molecular & Translational Medicine
Mike Breen Extravascular B Cell Populations in the Influenza A Virus Experienced Lung Mentors: Rachel Fearns and Joseph P. Mizgerd Microbiology PhD
Elim Na The cytoprotective effects of leukemia inhibitory factor during bacterial pneumonia Mentor: Lee J. Quinton Molecular and Translational Medicine PhD
Christine Odom Liver-dependent lung remodeling during systemic inflammation alters responses to secondary infection Mentor: Lee J. Quinton Microbiology PhD
Carolina Lyon De Ana Novel subsets of resident lymphocytes in murine lungs recovered from pneumococcal pneumonia Mentor: Joseph P. Mizgerd Microbiology PhD
Emad Arafa Recovery from pneumococcal pneumonia remodels the pool of alveolar macrophages Mentor: Joseph P. Mizgerd Molecular and Translational Medicine PhD
Kim Barker The establishment and function of lung resident memory B cells after bacterial respiratory infection Mentor: Joseph P. Mizgerd Microbiology PhD
Elizabeth Becker Derivation of airway epithelium transcriptomic signatures of COPD phenotypes Mentor: Katrina Steiling Bioinformatics GRS PhD
Julia Hicks-Berthet Roles for YAP and TAZ in Lung Epithelial Biology Mentor: Bob Varelas Biochemistry PhD
Xu Ke Airway Gene Expression Alterations in Association with Radiographic Abnormalities of the Lung Mentor: Marc Lenburg Bioinformatics MD/PhD
Nathan Kingston Roles for Hippo effectors TAZ and YAP in cancer and fibrosis Mentor: Bob Varelas Biomedical Sciences (PiBS) PhD
Xingyi Shi Bronchial Gene Expression Associated with Airway Premalignancy and Lung Cancer Subtypes Mentor: Marc Lenburg Bioinformatics PhD
Yuliang (Leon) Sun The role of ATP binding cassette A3 (ABCA3) in health and disease using pluripotent stem cell-derived type II alveolar epithelial cells Mentor: Darrell Kotton Molecular and Translational Medicine PhD
Jiarui Zhang Genomic biomarker development to impact clinical management of patients at risk for lung cancer Mentor: Marc Lenburg Molecular and Translational Medicine PhD
Anant Balijepalli Design, synthesis, characterization, and evaluation of a cationic poly-amido-saccharide towards biocompatible nucleic acid delivery Mentor: Mark Grinstaff Biomedical Engineering PhD
Sean Corbett Approaches for identifying lung cell type responses to perturbation Mentor: Marc Lenburg Bioinformatics PhD
Yuri Kim Liver-dependent protection during pneumonia and sepsis Mentor: Lee J. Quinton Molecular and Translational Medicine PhD
George Kwong Induced pluripotent stem cell reporter systems for smooth muscle cell sheet engineering Mentor: Darrell Kotton Biomedical Engineering PhD
Timothy Norman Origin and maturation of the pulmonary lymphatic endothelium Mentor: Alan Fine Pathology PhD
Alicia Wooten Pneumococcal phosphodiesterase 2 mutation elicits a unique type I interferon response in macrophages Mentor: Joseph P. Mizgerd Molecular and Translational Medicine PhD
Julia Barrios Neural regulation of the pulmonary neuroendocrine system induce mucus overproduction Mentor: Xingbin Ai Molecular and Translational Medicine PhD
Grant Duclos Characterization of smoking-associated transcriptomic alterations to the human bronchial epithelium Mentor: Avrum Spira Molecular and Translational Medicine PhD
Terry Hsieh Mild traumatic brain injury augments innate immune responses through neurokinin and cholinergic signaling Mentor: Daniel Remick Pathology Md/PhD
Anjali Jacob Generation of mature type II alveolar epithelial cells from human pluripotent stem cells Mentor: Darrell Kotton Molecular and Translational Medicine PhD
Jacob Josef Kantrowitz Transcriptomic alterations underlying pathogenesis and carcinogenesis in COPD Mentor: Marc Lenburg Molecular and Translational Medicine MD/PhD
Ana Brandusa Pavel Multi-omics data integration for the detection and characterization of smoking related lung diseases Mentor: Marc Lenburg Bioinformatics PhD
Katherine Benson McCauley Pluripotent stem cell modeling of airway epithelial fate Mentor: Darrell Kotton Molecular Biology PhD
Joseph Perez-Rogers Development of a minimally invasive molecular biomarker for early detection of lung cancer Mentor: Marc Lenburg/Avrum Spira Bioinformatics PhD
Aleksander Szymaniak Polarity and Hippo signaling in epithelial cell fate regulation Mentor: Bob Varelas Biochemistry PhD
Fadie Coleman: Influence of macrophage NF-kappaB activation on pneumococcal pneumonia Mentor: Joseph P. Mizgerd Microbiology PhD
Elyse Kozlowski: Functional roles for the terminal uridyltransferase enzymes Zcchc6 and Zcchc11 in mammalian biology Mentor: Matthew R. Jones Molecular and Translational Medicine PhD
Nicole Stauffer Smith: Respiratory infections with pneumococci establish multi-pronged heterotypic protection against pneumonia Mentor: Joseph P. Mizgerd Pathology and Laboratory Medicine PhD
Greg Wasserman: A discrete population of ciliated cells express the piRNA binding protein MIWI2 to regulate lung inflammation Mentor: Matthew R. Jones Microbiology PhD
Kristie Hilliard The functional significance of the lung-liver axis during pneumonia Mentor: Lee J. Quinton Microbiology PhD
Rebecca Kusko Integrative transcriptomics in smoking related lung diseases Mentor: Avrum Spira Genetics & Genomics PhD
Teresa Wang Transcriptomics of the human airway epithelium reflect the physiologic response to inhaled environmental pollutants Mentor: Marc Lenburg Bioinformatics PhD
Joseph Gerrein Using gene and microRNA expression in the human airway for lung cancer diagnosis Mentor: Avrum Spira Bioinformatics PhD
Kahkeshan Hijazi The airway transcriptome as a measure of injury response to and recovery from smoking and alternative tobacco products Mentor: Marc Lenburg Bioinformatics PhD
John Mahoney The role of Yap in lung development Mentor: Wellington Cardoso Pathology & Laboratory Medicine PhD
Linh Aven Innervation defects as a mechanism of childhood asthma Mentor: Xingbin Ai Molecular Medicine, Cell and Molecular Biology PhD
John Brothers Characterizing and reassembling the COPD and ILD transcriptome using RNA-Seq Mentor: Marc Lenburg Bioinformatics PhD
Radhika Dixit The contribution of mesothelial cells to lung development Mentor: Alan Fine Molecular Medicine PhD
Joshua Campbell Genome-wide characterization of microRNA and gene expression patterns in smoking-related lung disease Mentor: Marc Lenburg Bioinformatics PhD
Julie Gil Zeskind Gene expression alterations associated with progression of emphysema and small airway disease in smokers with COPD Mentor: Avrum Spira/Marc Lenburg Bioinformatics PhD
Adam Gower Discovering biological connections between experimental conditions based on common patterns of differential gene expression Mentor: Avrum Spira/Marc Lenburg Bioinformatics PhD
MIT Libraries home DSpace@MIT
- DSpace@MIT Home
- MIT Libraries
- Graduate Theses
Identifying prognostic biomarkers for cancer using gene expression data

Other Contributors
Terms of use, description, date issued, collections.
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 12 March 2021
A highly expressed mRNA signature for predicting survival in patients with stage I/II non-small-cell lung cancer after operation
- Meiling Yang 1 ,
- Meihua Li 1 na1 &
- Zhiyi He 1 na1
Scientific Reports volume 11 , Article number: 5855 ( 2021 ) Cite this article
2323 Accesses
4 Citations
2 Altmetric
Metrics details
- Cancer genomics
- Gene expression
- Genetic markers
There is an urgent need to identify novel biomarkers that predict the prognosis of patients with NSCLC. In this study,we aim to find out mRNA signature closely related to the prognosis of NSCLC by new algorithm of bioinformatics. Identification of highly expressed mRNA in stage I/II patients with NSCLC was performed with the “Limma” package of R software. Survival analysis of patients with different mRNA expression levels was subsequently calculated by Cox regression analysis, and a multi-RNA signature was obtained by using the training set. Kaplan–Meier estimator, log-rank test and receiver operating characteristic (ROC) curves were used to analyse the predictive ability of the multi-RNA signature. RT-PCR used to verify the expression of the multi-RNA signature, and Westernblot used to verify the expression of proteins related to the multi-RNA signature. We identified fifteen survival-related mRNAs in the training set and classified the patients as high risk or low risk. NSCLC patients with low risk scores had longer disease-free survival than patients with high risk scores. The fifteen-mRNA signature was an independent prognostic factor, as shown by the ROC curve. ROC curve also showed that the combined model of the fifteen-mRNA signature and tumour stage had higher precision than stage alone. The expression of fifteen mRNAs and related proteins were higher in stage II NSCLC than in stage I NSCLC. Multi-gene expression profiles provide a moderate prognostic tool for NSCLC patients with stage I/II disease.
Similar content being viewed by others
An eleven-gene risk model associated with lymph node metastasis predicts overall survival in lung adenocarcinoma

Identification of molecular subtypes and a prognostic signature based on m6A/m5C/m1A-related genes in lung adenocarcinoma
A novel 14-gene signature for overall survival in lung adenocarcinoma based on the Bayesian hierarchical Cox proportional hazards model
Introduction.
Non-small-cell lung cancer (NSCLC) is a disease with high morbidity and mortality rates, accounting for approximately 85% of lung cancer cases 1 , 2 . Although Surgery plays a pivotal role in treating NSCLC, 5-year survival rates of NSCLC after surgical resection are commonly accepted to be 60% to 80% for stage I and 30% to 50% for stage II 3 . The prognosis is more favourable in localized or limited advanced stages. The risk of recurrence peaks within the first 2 years after the operation 4 . Most postoperative recurrences are found during routine follow-up when patients are asymptomatic. Hence, it is urgent to explore effective NSCLC prognostic biomarkers to help optimize clinical management and ultimately further improve clinical outcome.
Gene expression can be used as a surrogate measurement of cancer disease phenotype 5 , 6 . Multiple gene signatures are found by using bioinformatics technology, and considered to have an intimate association with the prognosis of NSCLC 7 , 8 , 9 . High expression of some genes is closely related to cancer progression, which can be used to determine patient prognosis 10 , 11 , 12 . As such, numerous highly expressed genes with various inherent and acquired genetic alterations have been shown to influence NSCLC prognosis 1 , 7 , 8 , 13 , 14 . Screening tumor markers based on bioinformatics technology is a hot spot in current research, so the aim of this study is try to use a new algorithm to screen highly expressed mRNAs may have significant prognostic value in the recurrence of patients with NSCLC.
Microarray technology and bioinformatic analysis have been increasingly regarded as useful methods to identify biomarkers as diagnostic and prognostic tools 15 , 16 . With the help of gene expression databases such as Gene Expression Omnibus (GEO), it is easy to obtain abundant expression data for NSCLC. It is very helpful to analyse NSCLC at the genetic level. These resources have improved our ability analyse NSCLC at the genetic level. Data on individual patients' mRNA profiles and clinical information can be obtained from Affymetrix human genome U133 plus 2.0 array 17 , 18 and GEO data sets, after screening the data of mRNA, stage I and II patients are complete and suitable for analysis using bioinformatics, so we performed this research to find a multi-RNA prognostic signature of highly expressed mRNAs for predicting relapse in stage I/II patients with NSCLC after surgery by analysing the GEO data.
Identify survival-related mRNA in the training set
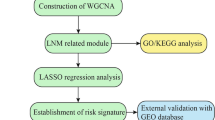
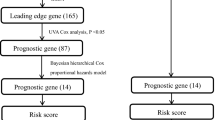
The fifteen-mRNA signature of highly expressed mRNAs associated with survival was developed and validated as shown in Fig. 1 . We identified highly expressed mRNAs from GSE31210 by using microarray data. Differentially expressed mRNAs were selected by volcano plot filtering (fold change ≥ 1 and P -value ≤ 0.05, Fig. 2 ). The relationship between RFS, survival state and high expression genes in NSCLC patients was performed using univariate Cox regression analysis in the training set, "risk score" of highly expressed genes in NSCLC prognosis was calculated. The higher the risk score, the greater the correlation between mRNA and RFS. According to the results, the fifteen-mRNA signature was significantly associated with RFS (n = 226, GSE31210; Table 1 ). UBE2F, TMSB10 and GAPDH were negative coefficients, indicating that patients with higher levels of expression had better outcomes than patients with lower levels of expression. Twelve mRNAs (UBC, TUBA1B, PPIA, PML, PKM, MESDC2, LDHA, HMOX1, FGFR1OP, CFB, ALDOA and ADAM10) were considered to be positive coefficients, so high levels of these mRNAs were associated with worse outcomes. Heatmap visualized distributions of fifteen-mRNA and risk scores in the training set and two independent GEO cohorts (Fig. 3 ).
Development and validation of the fifteen-mRNA signature shown as study flow.
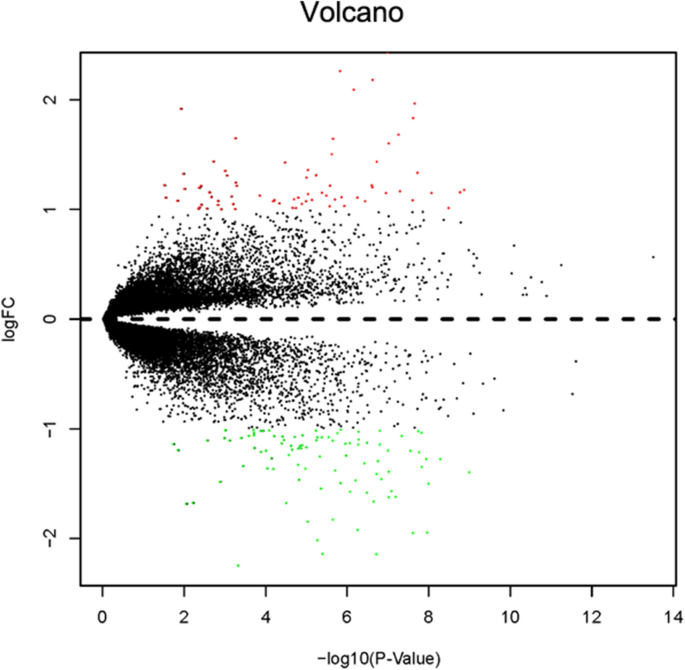
Volcano plot of mRNA expression in the training set (n = 226, GSE31210). The red points in the volcano plot represents the statistically significant highly expressed mRNAs, and the green points represent mRNAs with significantly low expression.
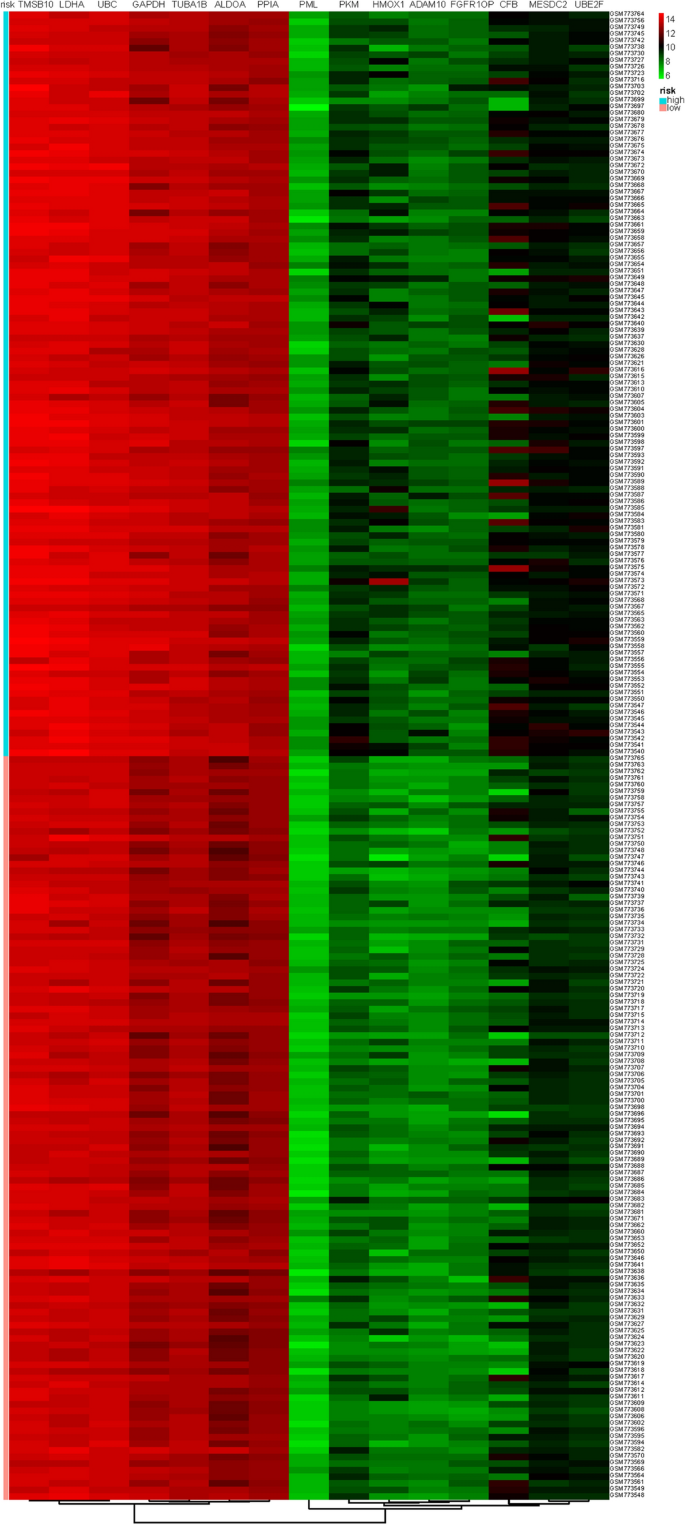
Heatmap of the fifteen-mRNA signature and risk scores in the training set(n = 226, GSE31210). The “pheatmap” package of R software (version 3.5.1) was used to generate the heatmap.
Survival analyses between low-risk and high-risk groups
On the basis of the expression of mRNAs and their regression coefficients in the multivariate Cox model, we determined individual patient risk scores according to the fifteen-mRNA in the training set, internal validation set and external validation set. As the median value was used as the cutoff value, NSCLC patients in each set were classified into the low-risk group or the high-risk group. Figure 4 shows the distributions of risk score and RFS status in each set, it demonstrated NSCLC patients who had high risk scores had a higher risk of relapse after surgery. The clinical characteristics of low-risk group and high-risk group patients in these three sets are shown in Table 2 . As our result, the clinical characteristics of the external independent variables (age, sex, stage) between the low-risk group and the high-risk group were not significantly different. RFS analyses were performed by log-rank test to determine the differences between high-risk and low-risk groups in these three sets (Fig. 5 and Table 3 ); lower scores were associated with longer RFS, and higher scores were associated with shorter RFS in each set ( P < 0.05). These results suggest that these fifteen-mRNA can distinguish NSCLC patients with different prognosis, and can be used in subsequent studies.
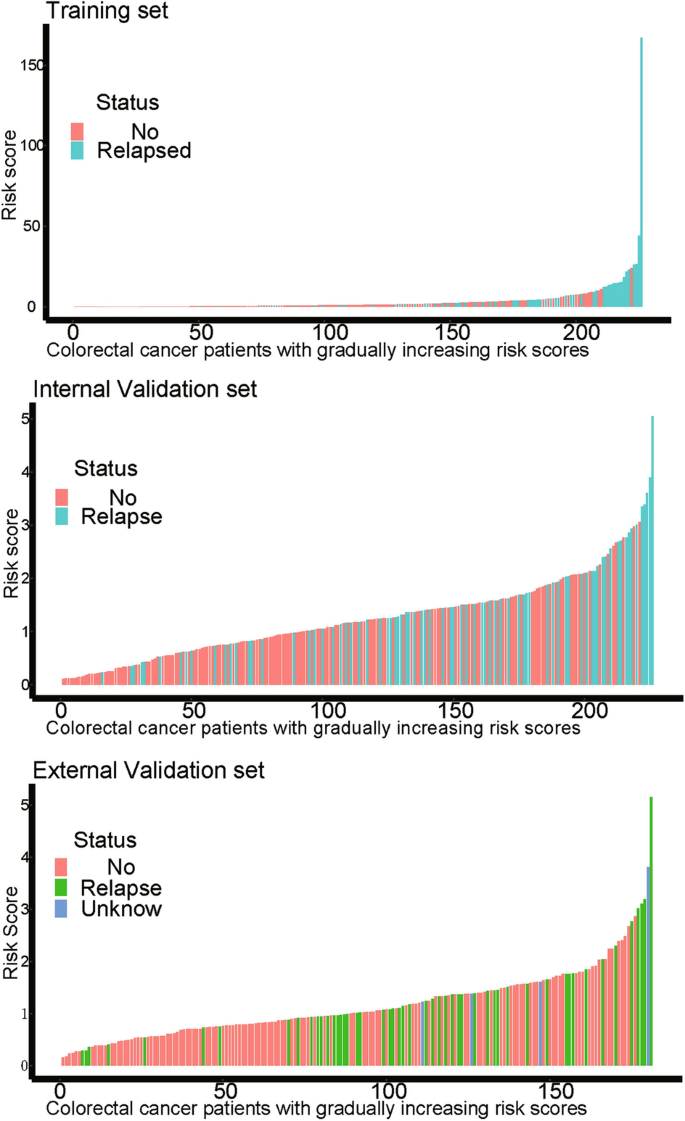
The distributions of RFS status and risk score in the training (n = 226, GSE31210) internal validation (n = 226, GSE30219) and external validation (n = 181, GSE50081) sets. The results showed that patients with recurrent NSCLC had a high risk score. Abbreviation: RFS, disease-free survival.
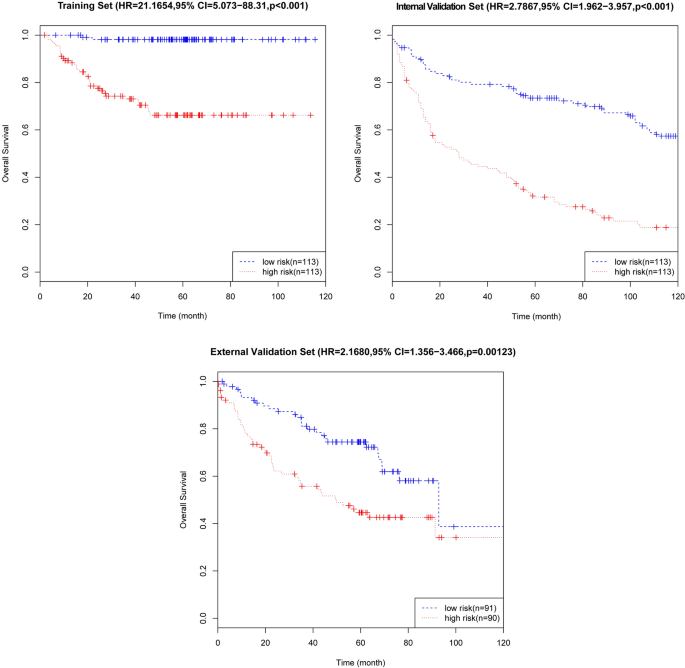
Kaplan–Meier curves of disease-free survival on the basis of the fifteen-mRNA signature in the training, internal validation, and external validation sets. As the result denmonstrated lower scores were associated with longer RFS, and higher scores were associated with shorter RFS in each set ( P < 0.05).
Multivariate Cox regression analysis of the fifteen-mRNA signature and clinical information in each set
The relationship of the fifteen-mRNA signature, clinical information (sex, age, stage) and RFS in each set was analysed by multivariate Cox regression analysis (Table 4 ). As our data showed, the fifteen-mRNA signature was significantly related to the RFS as well as clinical characteristic in these datasets of NSCLC patients (all P < 0.05).
ROC analysis of the fifteen-mRNA signature and stage in each set
The area under the curve (AUC) of the ROC curve was used to analyse the RFS of fifteen-mRNA signatures and stages in each set (Fig. 6 ). As the figure shows, the AUC of the fifteen-mRNA signature was higher than that of stage alone in the training ( P < 0.05) and internal validation sets ( P < 0.05). The combined model’s (fifteen-mRNA signature and tumour stage) AUC was higher than that of stage alone in each set ( P < 0.05). It suggests that fifteen-mRNA signatures have good predictive power to the prognosis of NSCLC patients.
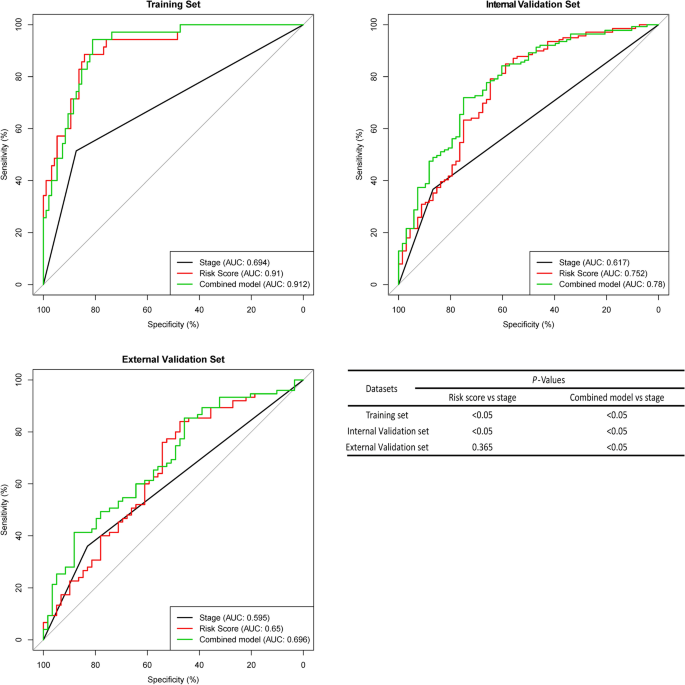
ROC curves of the combined model of the fifteen-mRNA signature and stage, the fifteen-mRNA signature and stage alone for each set. The AUC of the fifteen-mRNA signature was higher than that of stage alone in the training ( P < 0.05) and internal validation sets ( P < 0.05). The combined model’s (fifteen-mRNA signature and tumour stage) AUC was higher than that of stage alone in each set ( P < 0.05). Abbreviation: AUC, area under the curve.
Comparison of RFS in the combined set (training set and internal validation set)
To further verify the efficacy of this fifteen-mRNA signature, we merged the training set and internal validation set into a combined set (n = 407, GSE31210 and GSE30219) and compared the RFS of the low-risk group (n = 204) and high-risk group (n = 203). The results show that the RFS of the high-risk group was significantly shorter than that of the low-risk group ( P < 0.001, Table 5 ). Survival analysis of clinical information (sex, age, stage) and mRNA in the combined set using multivariate Cox regression. The results showed a significant correlation between our mRNA signature and RFS (HR = 2.30743, 95% CI = 1.7407–3.059, P < 0.001; Table 6 ). The Kaplan–Meier curve further showed that the RFS of the high-risk group was significantly shorter than that of the low-risk group ( P < 0.001, Fig. 7 ). The results in combined set are consistent with our previous analysis.
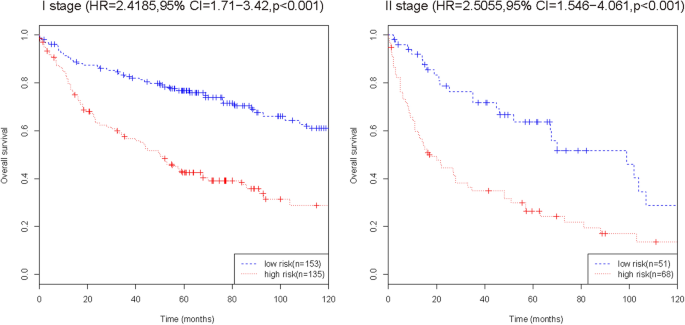
Kaplan–Meier curve analysis of RFS according to the fifteen-mRNA signature for stage I/II patients in the combined training set and internal validation set. The Kaplan–Meier curve further showed that the RFS of the high-risk group was significantly shorter than that of the low-risk group ( P < 0.001).
GO and KEGG functional enrichment analysis
We used GO and KEGG enrichment to identify the biological functions and signalling pathways of fifteen-mRNA signature. The results showed that the fifteen-mRNA signature was significantly associated with 94 GO terms (Fig. 8 ) and 20 KEGG pathways (Fig. 9 ). The GO terms mainly fit into three functional categories: carboxylic acid biosynthetic process (GO: 0,046,394), coenzyme metabolic process (GO: 0,006,732), and purine ribonucleoside triphosphate metabolic process (GO: 0,009,205). Glycolysis/gluconeogenesis (KEGG: 00010) and HIF-1 signalling pathway (KEGG: 04,066) were the main KEGG pathways involved.
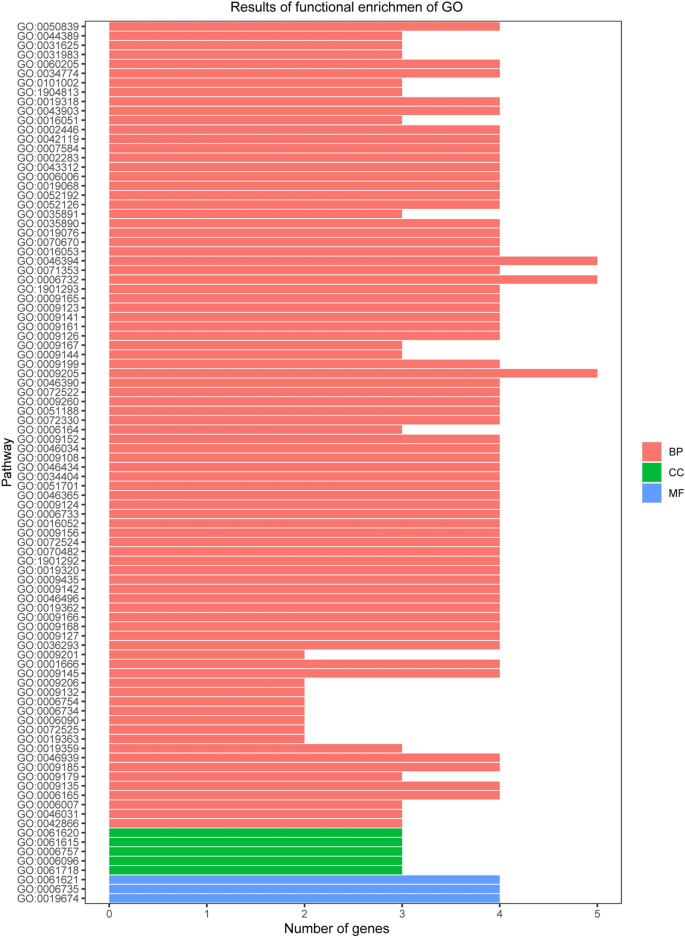
Functional enrichment analysis by GO category (BP: biological process; CC: cell component; MF: molecular function). The GO terms mainly fit into three functional categories: carboxylic acid biosynthetic process (GO: 0,046,394), coenzyme metabolic process (GO: 0,006,732), and purine ribonucleoside triphosphate metabolic process (GO: 0,009,205).
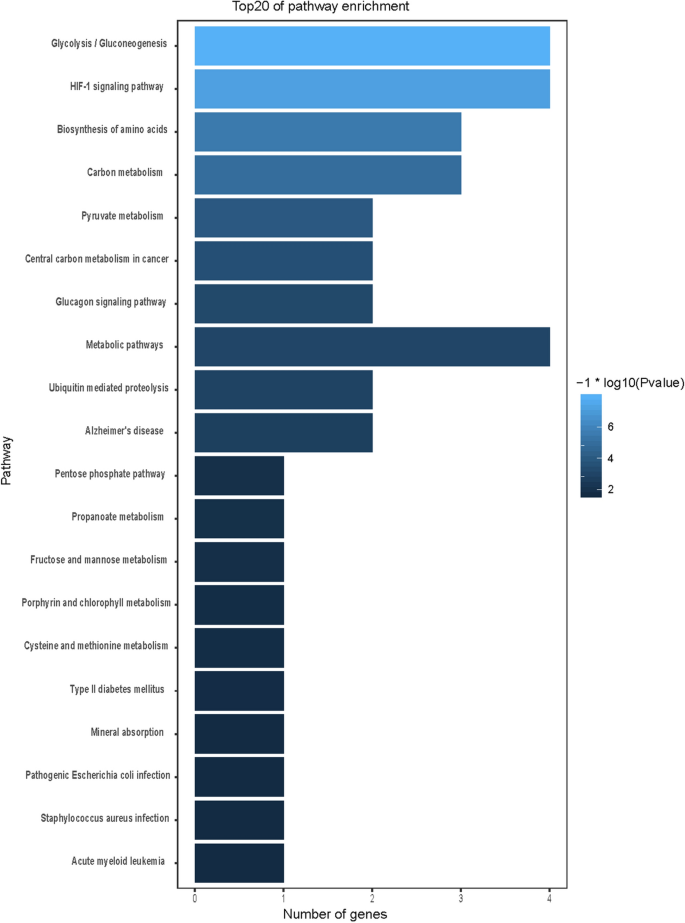
Calculated results of KEGG functional enrichment. Glycolysis/gluconeogenesis (KEGG: 00010) and HIF-1 signalling pathway (KEGG: 04,066) were the main KEGG pathways involved.
Protein–protein interaction analysis and mRNA expression validation
STRING online software was used to analyse the interaction between proteins encoded by the fifteen mRNAs (Fig. 10 A), and key genes were analysed according to the number of nodes using R software (version 3.5.1). Nodes were mainly interrelated with GAPDH and UBC, so these two proteins were speculated to be the key proteins in this protein–protein interaction network (Fig. 10 B). GEPIA online software was used to verify the expression of the fifteen mRNAs in stage I and II patients with lung adenocarcinoma and lung squamous cell carcinoma. The expression of ALDOA, CFB, GAPDH, LDHA, MESDC2, PPIA, TMSB10, TUBA1B, and UBE2F was higher in stage II patients than in stage I patients (Fig. 11 , P < 0.05).
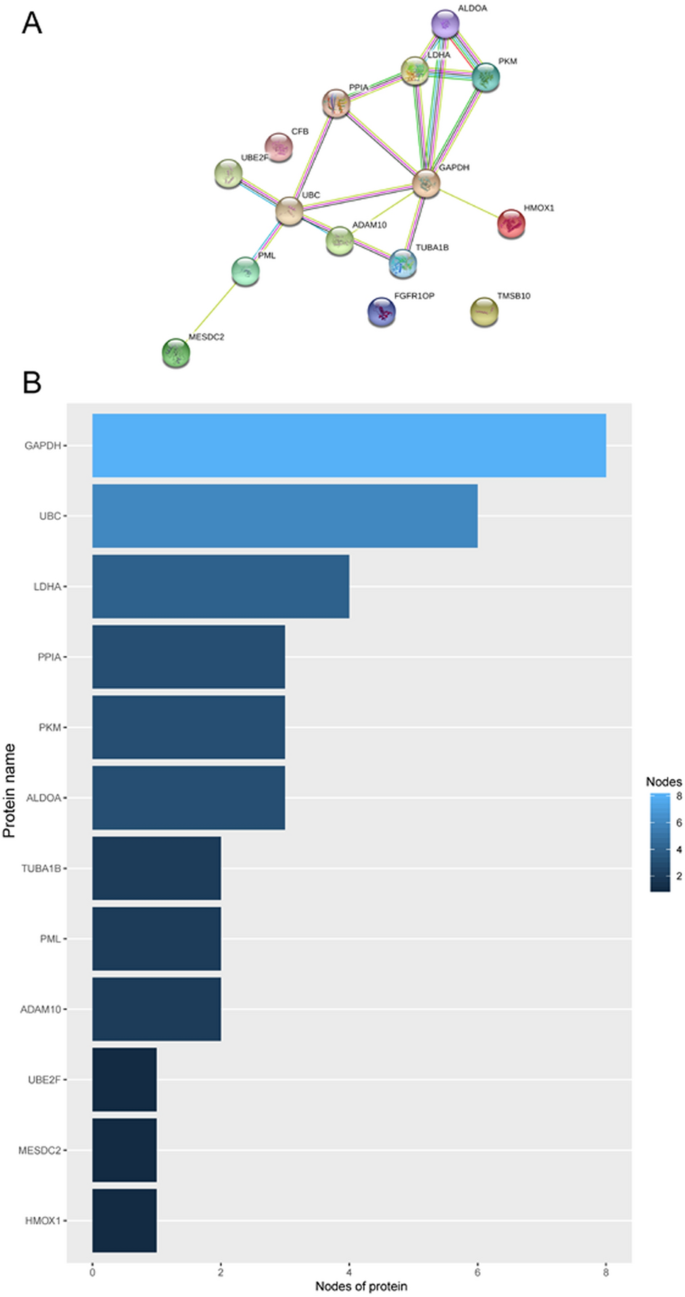
Protein–protein interaction network ( A ) and nodes ( B ) of proteins encoded by the fifteen mRNAs in the signature.
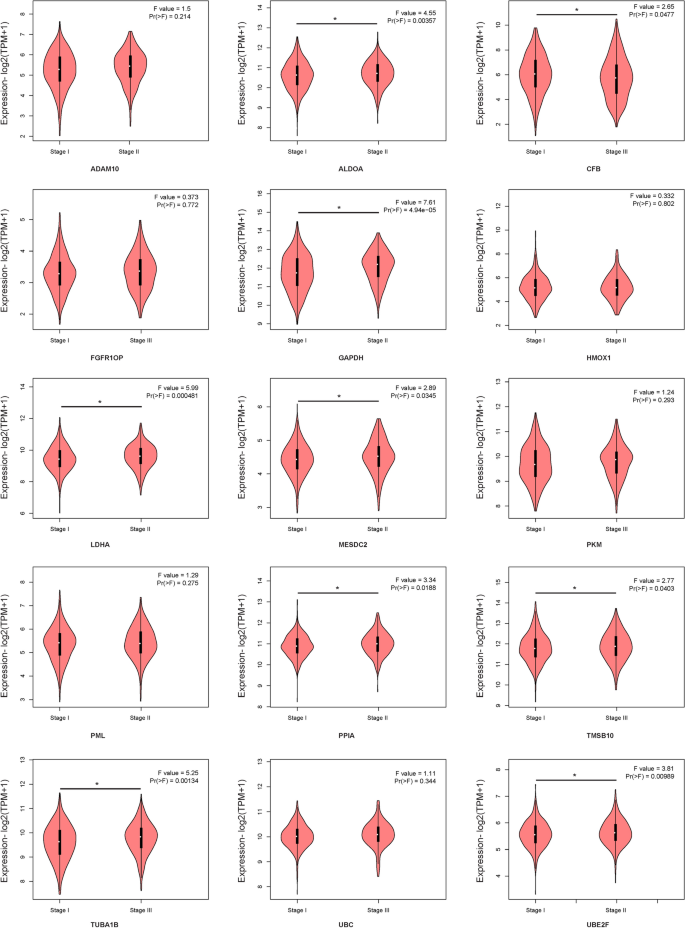
GEPIA online software verified the expression of the fifteen-mRNA signature in stage I/II patients with lung adenocarcinoma and lung squamous cell carcinoma.
The fifteen-mRNA mRNA expression in patients with NSCLC
We used PCR to verify the expression of the fifteen-mRNA in lung cancer tissues of NSCLC patients, and the results showed that the fifteen-mRNA was significantly higher in stage II NSCLC than in stage I (Fig. 12 , P < 0.05).
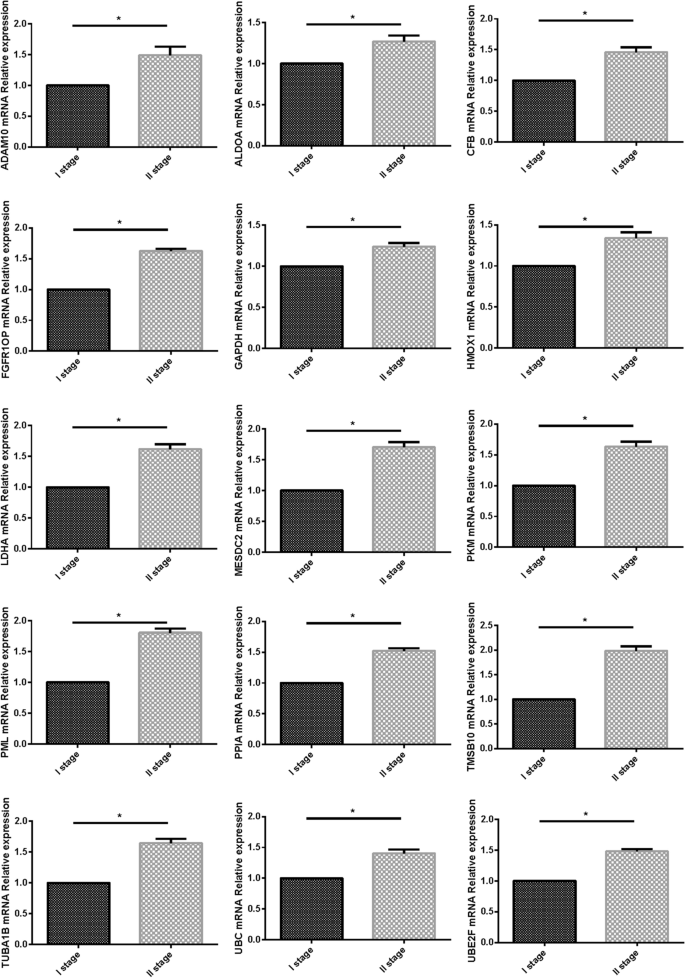
The expression of 15 mRNAs detected by RT-PCR. 15 mRNAs was significantly higher in stage II NSCLC than in stage I ( P < 0.05).
The proteins related to fifteen-mRNA mRNA expression in patients with NSCLC
We used westernblot to verify the expression of proteins related to the fifteen-mRNA in lung cancer tissues of NSCLC patients, and the results showed that these proteins was significantly higher in stage II NSCLC than in stage I (Fig. 13 , P < 0.05).
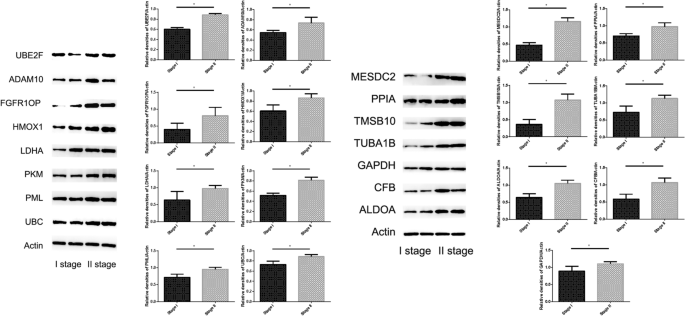
The expression of 15 mRNAs related protein in tumor tissue detected by Westernblot. These 15 mRNAs related protein was significantly higher in stage II NSCLC than in stage I ( P < 0.05).
In general, the TNM system is a widely used staging system among clinicians 19 , 20 , and TNM staging is essential for evaluating outcomes in clinical practice and for providing some indication of prognosis for survival 21 . Unfortunately, current methods of classification and staging for NSCLC are not completely reliable or sufficiently precise 22 , 23 , 24 . The progression and prognosis of tumours are related to the high expression of some genes 25 , 26 . The aim of this study was to characterize tumour recurrence and analyse genes related to the increased risk of recurrence in NSCLC. Bioinformatics analysis is currently considered to be an important tool for identifying tumour biomarkers. We profiled NSCLC mRNA by analysing the microarray data of the Affymetrix human genome U133 plus 2.0 array downloaded from GEO. High mRNA expression in stage I/II NSCLC was determined by the “Limma” package in the training set (GSE31210). Univariate Cox proportional hazards regression was used to analyse relationship between high expression genes and patient’s survival time and prognosis in NSCLC, the "risk score" of highly expressed genes in NSCLC prognosis was calculated, In this algorithm, the high risk socre of mRNA related with the poor prognosis of NSCLC. We selected 15 mRNAs with the highest risk score through this algorithm, and peculate that these 15 mRNAs are closely related to the prognosis of NSCLC. In order to study the predictive ability of these 15 mRNAs, we verify them in the training set and two independent GEO cohorts (GSE30219 and GSE50081), and the mRNA signature showed prognostic significance in three cohorts. Many factors, such as sex, age, and stage, are thought to be possible pathogenesis of NSCLC cancer. We analysed NSCLC patient RFS by multivariate Cox regression, and our results showed that the mRNA signature was associated with patient RFS. The mRNA signature performed better than stage alone, and the combined use of RNA and stage performed the best. The combined set, considering the mRNA signature and stage, was significantly associated with patient RFS; in the same stage, our mRNA signature was still significantly associated with patient RFS, and patients with low risk scores had significantly longer RFS. The fifteen-mRNA classifier has a very high HR and a very broad CI in the training set compared with the two other sets, we speculate it may be the instability of the training set. Furthermore, the ROC curve shownd that AUC of the fifteen-mRNA signature was higher than that of stage alone in the training and internal validation sets. The combined model’s (fifteen-mRNA signature and tumour stage) AUC was higher than that of stage alone in each set. Results of ROC curve suggests these fifteen-mRNA signatures as an independent prognostic factor in NSCLC. Finally, our bioinformatics analysis results shown our fifteen-mRNA signature is a novel biomarker with useful applications in predicting NSCLC prognosis.
To determine the biological relationship and signalling pathways among the fifteen mRNAs in the signature, we performed GO and KEGG analyses. Functional categories of the fifteen mRNAs were mainly involved in three GO terms, including the carboxylic acid biosynthetic process (GO: 0,046,394), coenzyme metabolic process (GO: 0,006,732), and purine ribonucleoside triphosphate metabolic process (GO: 0,009,205). All three pathways are considered to be closely related to tumours 27 , 28 , 29 , 30 . The main KEGG pathways involved included glycolysis/gluconeogenesis (KEGG: 00010) and the HIF-1 signalling pathway (KEGG: 04,066). Glycolysis is a universal pathway in living cells, and the glycolysis rate is 200 times higher in tumour cells than in normal cells 31 . Previous studies have shown that inhibition of HIF-1 represents a novel approach to cancer therapy 32 , 33 . We analysed the protein–protein interactions between proteins encoded by fifteen mRNAs. GAPDH and UBC were speculated to be the key proteins in this protein–protein interaction network according to their nodes, it suggest these two mRNA may play the key role of 15 mRNA. The expression of the fifteen mRNAs was validated by GEPIA online software, and the expression levels of ALDOA, CFB, GAPDH, LDHA, MESDC2, PPIA, TMSB10, TUBA1B, and UBE2F were higher in stage II patients than in stage I patients with lung adenocarcinoma and lung squamous cell carcinoma, which was consistent with our previous results in the gene sets.
Furthermore, we verified the expression of mRNA in NSCLC tumor tissues by RT-PCR and confirmed the expression of 15 mRNA related proteins by Westernblot. Our results showed that the expression of 15 mRNA genes was higher in stage II NSCLC than in stage I NSCLC, and the expression of 15 mRNA gene related proteins also showed the same situation, that is, in stage II is higher than in stage I. The fifteen-mRNA signature included twelve risky genes (UBC, TUBA1B, PPIA, PML, PKM, MESDC2, LDHA, HMOX1, FGFR1OP, CFB, ALDOA and ADAM10) and three protective genes (UBE2F, TMSB10 and GAPDH). Previous research showed that high tissue levels of PKM 34 , 35 , LDHA 36 , 37 , HMOX1 38 , FGFR1OP 39 , ADAM10 10 , 40 , ALDOA 41 , 42 , and GAPDH 43 , 44 were correlated with an increased risk of relapse in NSCLC patients. Low expression of UBC inhibits radiostasis and proliferation of NSCLC tumor cells 45 , UBE2F high expression promotes lung cancer cell survival 46 . CFB promote migration and proliferation of Cutaneous Squamous Cell Carcinoma 47 , and overexpression of TMSB10 relate with hepatocellular carcinoma and renal cell carcinoma 48 , 49 . Although there are no studies on eight of the mRNAs (TUBA1B, UBC, PPIA, PML, MESDC2, CFB, UBE2F, TMSB10) in prognosis of NSCLC, our experimental results shown these 15 mRNAs are involved in the progression of NSCLC, these experimental results provide evidence for the roles of these mRNAs in NSCLC and identify them as biomarkers.
The innovation of this research is identified mRNAs significantly related to RFS of NSCLC with a risk score via univariate Cox analysis, these 15 mRNAs have shown good predictive ability in the training set, internal validation set and external validation set. However, there were several limitations to our study. For example, further experiments are required to verify the clinical value of the signature. Limited by the clinical information of GEO data sets, we cannot identify the resection status of patients with NSCLC. Additionally, our experimental sample size is small, larger clinical trials may lead to more convincing results.
Our findings demonstrate a multiple-mRNA signature closely relate with tumour prognosis in stage I/II patients with NSCLC. It may aid in the development of novel biomarkers of NSCLC and offer new insights into NSCLC prognosis and may provide a new method for analyzing NSCLC based on Cox analysis.
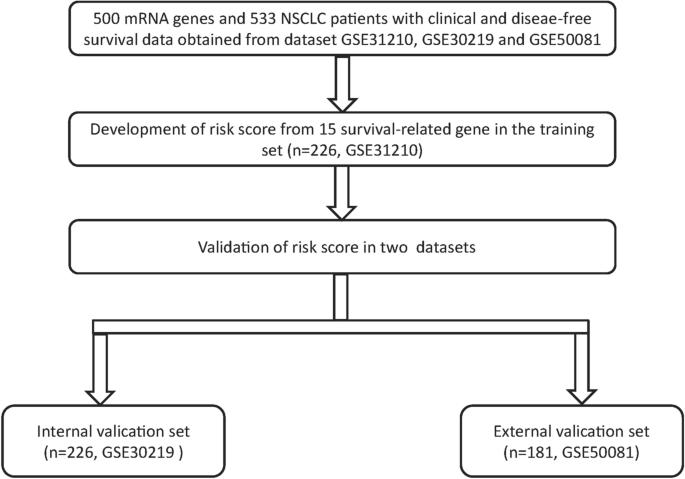
Data of NSCLC
Raw microarray data from all data sets were analysed using the Affymetrix human genome U133 plus 2.0 array (GSE31210, GSE30219 and GSE50081), the mRNA expression data were log2 transformed before statistical analysis, and the median value was used when multiple probes existed for a single target. There was a total of 627 stage I/II patients with NSCLC after excluding patients without Recurrence Free Survival(RFS) or clinical data, including 226 from GSE31210, 226 from GSE30219 and 181 from GSE50081. The 226 patients from GSE31210 were used as a training set, 226 patients from GSE30219 were used as an internal validation set, and 181 patients from GSE50081 were used as the external validation set. The training set was used to optimize the parameters of model, and the internal validation set was used to tune hyper-parameters to optimize the model, external validation set use for validating the robustness of the screening method to different data.
Tissue specimens
Tumor tissues were obtained from 8 patients underwent resection of NSCLC (mean age of 54.4 ± 2.3 years, six males). The samples were taken during thoracic surgery, all cancer tissues were identified by HE staining. Four patients were stage I and the rest were stage II, all patients with no history of COPD or other respiratory infectious diseases.
Real-time PCR (RT-PCR)
Total RNA was extracted from tumor tissues using the TRIzol reagent (TaKaRa, Dalian, China). The primer sequences of 15 mRNAs are listed in Table 7 . Qualitative and quantitative analysis of total RNA were using Nanodrop. RNA was reverse transcripted to cDNA and all samples carried out in triplicate and run RT-PCR on an ABI/PRISM 7500 according to the reagent manufacturer's instructions. RT-PCR was performed by SYBR Premix Ex TaqTM II (TaKaRa, Dalian, China).
Westernblot
After sufficiently ground and crushed tumor tissue, the protein in tumor tissue is extracted with radioimmunoprecipitation assay (RIPA) buffer, the expression level of 15 mRNAs related protein in cancer tissue were detected by Westernblot. The relevant antibodies used to detect the target protein are as follows: ADAM10 (dilution 1:1000; Abclone, Wuhan, Hubei, China), ALDOA (dilution 1:10,000; Abclone, Wuhan, Hubei, China), CFB (dilution 1: 1000; Abclone, Wuhan, Hubei, China), FGFR1OP (dilution 1: 1000; Abclone, Wuhan, Hubei, China), GAPDH (dilution 1:1000; Abclone, Wuhan, Hubei, China), HMOX1 (dilution 1:1000; Abclone, Wuhan, Hubei, China), LDHA (dilution 1:1000; Abclone, Wuhan, Hubei, China), MESDC2 (dilution 1:1000; Abclone, Wuhan, Hubei, China), PKM (dilution 1:1000; Abclone, Wuhan, Hubei, China), PML (dilution 1:1000; Abclone, Wuhan, Hubei, China), PPIA (dilution 1:1000; Abclone, Wuhan, Hubei, China), TMSB10 (dilution 1:1000; Sigma-Aldrich Chemicals, St. Louis, MO, USA)), TUBA1B (dilution 1:1000; Abclone, Wuhan, Hubei, China), UBC (dilution 1:1000; Abclone, Wuhan, Hubei, China), UBE2F (dilution 1:1000; Abclone, Wuhan, Hubei, China), Beta Actin (dilution 1:1000; Abclone, Wuhan, Hubei, China).
Statistical analysis
The “survival” package of R software (version 3.5.1) was used to perform survival analysis. Univariate Cox regression analysis was used to evaluate the association between the expression level of mRNA, NSCLC patients’ RFS and patients’survival state in the training set. mRNA expression was considered to be significantly different when the P -value was < 0.05, and multivariate Cox regression analysis of highly expressed mRNAs was used to calculate their risk score regression coefficients in the training set 50 , 51 , 52 . The median value of risk scores in the training set was used as the cutoff point, and NSCLC patients in the training, internal validation, and external validation sets were classified as low risk or high risk corresponding to the cutoff. The Kaplan–Meier estimator and log-rank test were used to assess survival differences between the two groups. Multivariate Cox regression analysis was used to compare the efficacy of the risk score system and the efficacy of clinical characteristics such as stage, age, and sex. ROC curves were used to show the predictive value of RFS in the combined model (risk score combined with stage), risk score model and stage alone. To generate the ROC curves, patients with NSCLC who had a duration of less than 5 years of RFS were excluded if they did not relapse at the last follow-up. We referred to the previous method, set 60 months as the cutoff value of RFS for reasearch the 5-year survival rates, and the remaining NSCLC patients were divided into two groups by this cutoff value 53 , 54 . The “pROC” package of R software was used to generate the ROC curve of RFS. Differences observed in the log-rank test, Cox regression analysis, and ROC analysis were considered to be significant if their P -values were < 0.05.
Results of RT-PCR and Westernblot are presented as means ± SD. Statistical analyses were calculated via SPSS (version 16.0.0; SPSS, Chicago, IL, USA). One-way ANOVA, Bonferroni post hoc correction (α = 0.0167), and Tukey test were conducted to evaluate significant differences in the data. Statistical significance was set at P < 0.05.
Functional enrichment analysis
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were based on the GeneCodis web tool ( http://genecodis.cnb.csic.es/ ) and KOBAS web tool ( http://kobas.cbi.pku.edu.cn/kobas3/?t=1 ) for functional enrichment analysis of these fifteen-mRNA signature. GO and KEGG category enrichments analyses had cutoff thresholds of P -value < 0.05. R software (version 3.5.1) was used to display significant enrichment results in graphical format.
STRING ( https://string-db.org/ ) online software was used to analyse the interaction between proteins of fifteen mRNAs and to screen key genes. Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/ ) online software was used to verify the expression of fifteen-mRNA in stage I and II patients with lung adenocarcinoma and lung squamous cell carcinoma.
Ethics approval and consent to participate
All samples were obtained with informed consent and all protocols were approved by the First Affiliated Hospital of Guangxi Medical University (Scientific and Research Ethics Committee, No. 2020(KY-E-142)). And written informed consent was obtained from all patients participated in our research. This study follows the ethical guidelines of the Declaration of Helsinki 1975.
Data availability
All data generated or analyzed during this study are included in this published article.
Abbreviations
Non-small cell lung cancer
Gene expression omnibus
Receiver operating characteristic
Gene ontology
Kyoto encyclopedia of genes and genomes
Gene expression profiling interactive analysis
Biological process
Cell component
Molecular function
Chen, H. Y. et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N. Engl. J. Med. 356 , 11–20. https://doi.org/10.1056/NEJMoa060096 (2007).
Article CAS PubMed Google Scholar
Cho, W. C. Application of proteomics in non-small-cell lung cancer. Expert Rev. Proteom. 13 , 1–4. https://doi.org/10.1586/14789450.2016.1121813 (2016).
Article CAS Google Scholar
Howington, J. A., Blum, M. G., Chang, A. C., Balekian, A. A. & Murthy, S. C. Treatment of stage I and II non-small cell lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 143 , e278S-e313S. https://doi.org/10.1378/chest.12-2359 (2013).
Hung, J. J. et al. Prognostic factors of postrecurrence survival in completely resected stage I non-small cell lung cancer with distant metastasis. Thorax 65 , 241–245. https://doi.org/10.1136/thx.2008.110825 (2010).
Article PubMed Google Scholar
Mansoori, B. et al. miR-142-3p as tumor suppressor miRNA in the regulation of tumorigenicity, invasion and migration of human breast cancer by targeting Bach-1 expression. J. Cell. Physiol. 234 , 9816–9825. https://doi.org/10.1002/jcp.27670 (2019).
Liu, T., Xu, Z., Ou, D., Liu, J. & Zhang, J. The miR-15a/16 gene cluster in human cancer: a systematic review. J. Cell. Physiol. 234 , 5496–5506. https://doi.org/10.1002/jcp.27342 (2019).
Tang, H. et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin. Cancer Res. 19 , 1577–1586. https://doi.org/10.1158/1078-0432.CCR-12-2321 (2013).
Article CAS PubMed PubMed Central Google Scholar
Robles, A. I. et al. An integrated prognostic classifier for stage I lung adenocarcinoma based on mRNA, microRNA, and DNA methylation biomarkers. J. Thorac. Oncol. 10 , 1037–1048. https://doi.org/10.1097/JTO.0000000000000560 (2015).
Xie, Y. et al. Validation of the 12-gene predictive signature for adjuvant chemotherapy response in lung cancer. Clin. Cancer Res. 25 , 150–157. https://doi.org/10.1158/1078-0432.CCR-17-2543 (2019).
Yoneyama, T. et al. ADAM10 sheddase activity is a potential lung-cancer biomarker. J. Cancer 9 , 2559–2570. https://doi.org/10.7150/jca.24601 (2018).
Zhou, H. et al. High expression of Toll-like receptor 5 correlates with better prognosis in non-small-cell lung cancer: an anti-tumor effect of TLR5 signaling in non-small cell lung cancer. J. Cancer Res. Clin. Oncol. 140 , 633–643. https://doi.org/10.1007/s00432-014-1616-4 (2014).
Ly, D., Zhu, C. Q., Cabanero, M., Tsao, M. S. & Zhang, L. Role for high-affinity IgE receptor in prognosis of lung adenocarcinoma patients. Cancer Immunol. Res. 5 , 821–829. https://doi.org/10.1158/2326-6066.CIR-16-0392 (2017).
Wang, M., Zhu, J., Lubman, D. M. & Gao, C. Aberrant glycosylation and cancer biomarker discovery: a promising and thorny journey. Clin. Chem. Lab. Med. 57 , 407–416. https://doi.org/10.1515/cclm-2018-0379 (2019).
Sun, R. et al. Metabolic gene NR4A1 as a potential therapeutic target for non-smoking female non-small cell lung cancer patients. Thorac. Cancer https://doi.org/10.1111/1759-7714.12989 (2019).
Article PubMed PubMed Central Google Scholar
Wu, Y. et al. Identification and characterization of sexual dimorphismlinked gene expression profile in hepatocellular carcinoma. Oncol. Rep. 42 , 937–952. https://doi.org/10.3892/or.2019.7217 (2019).
Wei, C. et al. Bioinformatics profiling utilized a nine immune-related long noncoding RNA signature as a prognostic target for pancreatic cancer. J. Cell. Biochem. 120 , 14916–14927. https://doi.org/10.1002/jcb.28754 (2019).
Riker, A. I. et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med. Genomics 1 , 13. https://doi.org/10.1186/1755-8794-1-13 (2008).
Hage-Sleiman, R. et al. Genomic alterations during p53-dependent apoptosis induced by gamma-irradiation of Molt-4 leukemia cells. PLoS ONE 12 , e0190221. https://doi.org/10.1371/journal.pone.0190221 (2017).
Roupret, M. et al. European guidelines for the diagnosis and management of upper urinary tract urothelial cell carcinomas: 2011 update. Eur. Urol. 59 , 584–594. https://doi.org/10.1016/j.eururo.2010.12.042 (2011).
Edge, S. B. & Compton, C. C. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann. Surg. Oncol. 17 , 1471–1474. https://doi.org/10.1245/s10434-010-0985-4 (2010).
Hattori, A., Takamochi, K., Okms, S. & Suzuki, K. New revisions and current in the eighth edition of the TNM classification for non-small cell lung cancer. Jpn. J. Clin. Oncol. 49 , 3–11. https://doi.org/10.1093/jjco/hyy142 (2019).
Van Bruwaene, S., Costello, A. J. & Van Poppel, H. Prognosis of node-positive bladder cancer in 2016. Minerva Urol. Nefrol. 68 , 125–137 (2016).
Google Scholar
Galon, J. et al. Towards the introduction of the “Immunoscore” in the classification of malignant tumours. J. Pathol. 232 , 199–209. https://doi.org/10.1002/path.4287 (2014).
Galon, J. et al. Cancer classification using the immunoscore: a worldwide task force. J. Transl. Med. 10 , 9. https://doi.org/10.1186/1479-5876-10-205 (2012).
Article Google Scholar
Ghasabi, M. et al. MicroRNAs in cancer drug resistance: basic evidence and clinical applications. J. Cell. Physiol. 234 , 2152–2168. https://doi.org/10.1002/jcp.26810 (2019).
Tian, W., Chen, J., He, H. & Deng, Y. MicroRNAs and drug resistance of breast cancer: basic evidence and clinical applications. Clin. Transl. Oncol. 15 , 335–342. https://doi.org/10.1007/s12094-012-0929-5 (2013).
Brea-Calvo, G., Rodriguez-Hernandez, A., Fernandez-Ayala, D. J., Navas, P. & Sanchez-Alcazar, J. A. Chemotherapy induces an increase in coenzyme Q10 levels in cancer cell lines. Free Radic. Biol. Med. 40 , 1293–1302. https://doi.org/10.1016/j.freeradbiomed.2005.11.014 (2006).
Jiang, Z., Woda, B. A. & Yang, X. M. J. alpha-Methylacyl coenzyme A racemase as a marker for prostate cancer. JAMA-J. Am. Med. Assoc. 287 , 3080–3081. https://doi.org/10.1001/jama.287.23.3080-a (2002).
Qian, Y., Wang, X., Li, Y., Cao, Y. & Chen, X. Extracellular ATP a new player in cancer metabolism: NSCLC cells internalize ATP in vitro and in vivo using multiple endocytic mechanisms. Mol. Cancer Res. MCR 14 , 1087–1096. https://doi.org/10.1158/1541-7786.MCR-16-0118 (2016).
Qian, Y. et al. Extracellular ATP is internalized by macropinocytosis and induces intracellular ATP increase and drug resistance in cancer cells. Cancer Lett. 351 , 242–251. https://doi.org/10.1016/j.canlet.2014.06.008 (2014).
Akram, M. Mini-review on glycolysis and cancer. J. Cancer Educ. 28 , 454–457. https://doi.org/10.1007/s13187-013-0486-9 (2013).
Mooring, S. R. & Wang, B. HIF-1 inhibitors as anti-cancer therapy. Sci. China Chem. 54 , 24–30. https://doi.org/10.1007/s11426-010-4187-5 (2011).
Semenza, G. L. HIF-1 and tumor progression: pathophysiology and therapeutics. Trends Mol. Med. 8 , S62–S67. https://doi.org/10.1016/s1471-4914(02)02317-1 (2002).
Ye, X. Y., Sun, Y. J., Xu, Y. H., Chen, Z. W. & Lu, S. Integrated In silico-in vitro discovery of lung cancer-related tumor pyruvate kinase M2 (PKM2) inhibitors. Med. Chem. 12 , 613–620. https://doi.org/10.2174/1573406412666160307151535 (2016).
Yuan, S. et al. Knockdown of the M2 isoform of pyruvate kinase (PKM2) with shRNA enhances the effect of docetaxel in human NSCLC cell lines in vitro. Yonsei Med. J. 57 , 1312–1323. https://doi.org/10.3349/ymj.2016.57.6.1312 (2016).
Danner, B. C. et al. Long-term survival is linked to serum LDH and partly to tumour LDH-5 in NSCLC. Anticancer Res. 30 , 1347–1351 (2010).
CAS PubMed Google Scholar
Nair, V. S., Gevaert, O., Davidzon, G., Plevritis, S. K. & West, R. NF-kappaB protein expression associates with (18)F-FDG PET tumor uptake in non-small cell lung cancer: a radiogenomics validation study to understand tumor metabolism. Lung Cancer 83 , 189–196. https://doi.org/10.1016/j.lungcan.2013.11.001 (2014).
Nitti, M. et al. HO-1 induction in cancer progression: a matter of cell adaptation. Antioxidants https://doi.org/10.3390/antiox6020029 (2017).
Mano, Y. et al. Fibroblast growth factor receptor 1 oncogene partner as a novel prognostic biomarker and therapeutic target for lung cancer. Cancer Sci. 98 , 1902–1913. https://doi.org/10.1111/j.1349-7006.2007.00610.x (2007).
Guo, J. et al. ADAM10 overexpression in human non-small cell lung cancer correlates with cell migration and invasion through the activation of the Notch1 signaling pathway. Oncol. Rep. 28 , 1709–1718. https://doi.org/10.3892/or.2012.2003 (2012).
Fu, H. et al. Aldolase A promotes proliferation and G1/S transition via the EGFR/MAPK pathway in non-small cell lung cancer. Cancer Commun. 38 , 18. https://doi.org/10.1186/s40880-018-0290-3 (2018).
Zhang, F. et al. Elevated transcriptional levels of aldolase A (ALDOA) associates with cell cycle-related genes in patients with NSCLC and several solid tumors. BioData Min. 10 , 6. https://doi.org/10.1186/s13040-016-0122-4 (2017).
Cuezva, J. M. et al. The bioenergetic signature of lung adenocarcinomas is a molecular marker of cancer diagnosis and prognosis. Carcinogenesis 25 , 1157–1163. https://doi.org/10.1093/carcin/bgh113 (2004).
Puzone, R. et al. Glyceraldehyde-3-phosphate dehydrogenase gene over expression correlates with poor prognosis in non small cell lung cancer patients. Mol. Cancer https://doi.org/10.1186/1476-4598-12-97 (2013).
Tang, Y. et al. Downregulation of ubiquitin inhibits the proliferation and radioresistance of non-small cell lung cancer cells in vitro and in vivo. Sci. Rep. 5 , 9476. https://doi.org/10.1038/srep09476 (2015).
Zhou, W. et al. Neddylation E2 UBE2F promotes the survival of lung cancer cells by activating CRL5 to degrade NOXA via the K11 linkage. Clin. Cancer Res. 23 , 1104–1116. https://doi.org/10.1158/1078-0432.CCR-16-1585 (2017).
Riihila, P. et al. Complement component C3 and complement factor B promote growth of cutaneous squamous cell carcinoma. Am. J. Pathol. 187 , 1186–1197. https://doi.org/10.1016/j.ajpath.2017.01.006 (2017).
Song, C., Su, Z. & Guo, J. Thymosin beta 10 is overexpressed and associated with unfavorable prognosis in hepatocellular carcinoma. Biosci. Rep. https://doi.org/10.1042/BSR20182355 (2019).
Xiao, R. et al. TMSB10 promotes migration and invasion of cancer cells and is a novel prognostic marker for renal cell carcinoma. Int. J. Clin. Exp. Pathol. 12 , 305–312 (2019).
CAS PubMed PubMed Central Google Scholar
Hu, Z. et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J. Clin. Oncol. 28 , 1721–1726. https://doi.org/10.1200/JCO.2009.24.9342 (2010).
Article ADS PubMed Google Scholar
Sun, G. et al. Identification of a five-gene signature with prognostic value in colorectal cancer. J. Cell. Physiol. 234 , 3829–3836. https://doi.org/10.1002/jcp.27154 (2019).
Wu, Y. S. et al. A four-miRNA signature as a novel biomarker for predicting survival in endometrial cancer. Gene 697 , 86–93. https://doi.org/10.1016/j.gene.2019.01.046 (2019).
Kang, J., D’Andrea, A. D. & Kozono, D. A DNA repair pathway-focused score for prediction of outcomes in ovarian cancer treated with platinum-based chemotherapy. J. Natl. Cancer Inst. 104 , 670–681. https://doi.org/10.1093/jnci/djs177 (2012).
Xu, G., Zhou, Y. & Zhou, F. Development and validation of an immunity-related classifier of nine chemokines for predicting recurrence in stage I-III patients with colorectal cancer after operation. Cancer Manag. Res. 10 , 4051–4064. https://doi.org/10.2147/CMAR.S174452 (2018).
Download references
Acknowledgements
Not applicable.
This work was supported by a grant from the National Natural Science Foundation of China (81860010).
Author information
These authors contributed equally: Meihua Li and Zhiyi He.
Authors and Affiliations
Department of Respiratory Medicine, The First Affiliated Hospital of GuangXi Medical University, Nanning, 530021, GuangXi, People’s Republic of China
Nan Ma, Lu Si, Meiling Yang, Meihua Li & Zhiyi He
You can also search for this author in PubMed Google Scholar
Contributions
N.M. and L.S. performed collected all the data and statistical analysis. N.M., L.S and M.Y. were responsible for biological experiment. Z.H. conceived of the study, and participated in its design and coordination. N.M. drafted the manuscript, which was revised by M.L. and Z.H. All authors read and approved the final manuscript.
Corresponding authors
Correspondence to Meihua Li or Zhiyi He .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Consent to publish
Additional information, publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary information., rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Ma, N., Si, L., Yang, M. et al. A highly expressed mRNA signature for predicting survival in patients with stage I/II non-small-cell lung cancer after operation. Sci Rep 11 , 5855 (2021). https://doi.org/10.1038/s41598-021-85246-x
Download citation
Received : 21 August 2020
Accepted : 24 February 2021
Published : 12 March 2021
DOI : https://doi.org/10.1038/s41598-021-85246-x
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
A novel chr1-mir-200 driven whole transcriptome signature shapes tumor immune microenvironment and predicts relapse in early-stage lung adenocarcinoma.
- Simon Garinet
- Audrey Didelot
- Hélène Blons
Journal of Translational Medicine (2023)
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Cancer newsletter — what matters in cancer research, free to your inbox weekly.
REVIEW article
Chimeric antigen receptor t-cell therapy in lung cancer: potential and challenges.

- 1 Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Department of Thoracic Surgery II, Peking University Cancer Hospital & Institute, Beijing, China
- 2 Department of Thoracic Surgery, The First Affiliated Hospital of Nanchang University, Nanchang, China
- 3 Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Laboratory of Biochemistry and Molecular Biology, Peking University Cancer Hospital & Institute, Beijing, China
Chimeric antigen receptor T (CAR-T) cell therapy has exhibited a substantial clinical response in hematological malignancies, including B-cell leukemia, lymphoma, and multiple myeloma. Therefore, the feasibility of using CAR-T cells to treat solid tumors is actively evaluated. Currently, multiple basic research projects and clinical trials are being conducted to treat lung cancer with CAR-T cell therapy. Although numerous advances in CAR-T cell therapy have been made in hematological tumors, the technology still entails considerable challenges in treating lung cancer, such as on−target, of−tumor toxicity, paucity of tumor-specific antigen targets, T cell exhaustion in the tumor microenvironment, and low infiltration level of immune cells into solid tumor niches, which are even more complicated than their application in hematological tumors. Thus, progress in the scientific understanding of tumor immunology and improvements in the manufacture of cell products are advancing the clinical translation of these important cellular immunotherapies. This review focused on the latest research progress of CAR-T cell therapy in lung cancer treatment and for the first time, demonstrated the underlying challenges and future engineering strategies for the clinical application of CAR-T cell therapy against lung cancer.
1 Introduction
Lung cancer is one of the most frequently occurring malignant tumors worldwide and is characterized by a substantially high malignancy and poor prognosis ( 1 ). According to the latest global cancer statistics, lung cancer remains the leading cause of cancer-related deaths worldwide ( 2 ). Lung cancer can be histologically classified into two main subtypes: small-cell lung carcinoma (SCLC) and non-small-cell lung carcinoma (NSCLC) ( 3 ). NSCLC accounts for approximately 85% of diagnosed lung cancer cases and can be further divided into adenocarcinoma, squamous cell carcinoma, and large cell carcinoma ( 4 , 5 ).
The present therapeutic measures for NSCLC primarily include surgical resection, chemoradiation, molecular-targeted therapy, and immunotherapy ( 6 ). The surgical resection procedure was based on the TNM stage of NSCLC patients. Conventional or stereotactic radiotherapy is applicable to patients with surgically unresectable NSCLC ( 7 ). Platinum-based double-agent combination chemotherapy is generally accepted as the standard chemotherapy regimen for NSCLC ( 8 ). Neoadjuvant chemotherapy is applied preoperatively to downgrade the cancer stage, whereas adjuvant chemotherapy is administered postoperatively, primarily involving cisplatin-based combination regimens ( 7 ). The primary molecular-targeted therapies include epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs), anti-EGFR monoclonal antibodies, fusion gene ALK and ROS1 inhibitors, and anti-vascular endothelial growth factor receptor monoclonal antibodies ( 9 – 12 ). Combined therapy with multiple immune checkpoint inhibitors, such as a combination of nivolumab and ipilimumab, has been shown to achieve better response rates than monotherapy ( 13 , 14 ).
Non-surgical treatment involving systemic chemotherapy plus radiotherapy is the mainstream procedure for SCLC patients because metastases occur when SCLC is newly diagnosed. Etoposide-platinum and topotecan are the standard first-line and second-line regimens for SCLC patients, respectively ( 15 , 16 ). Although SCLC is very sensitive to chemotherapy, many SCLC patients relapse due to the clinical development of chemoresistance. Moreover, nivolumab was the first FDA-approved immunotherapy agent for SCLC treatment ( 17 ). Several small molecular inhibitors, including PARP inhibitors, have also been demonstrated to exert anti-tumor activity in SCLC in clinical trials ( 18 , 19 ). However, due to the heterogeneity of tumors, it is imperative to explore effective novel therapies.
Chimeric antigen receptors (CARs) are engineered receptors that can enable modified T cells to recognize and kill tumor cells expressing a tumor-specific antigen ( 20 ). CAR-T cells contain two sections: autologous T cells separated from the peripheral blood of patients and integration of CARs into T cells through genetic engineering in the laboratory. Patient’s T cells are extracted, isolated, and genetically engineered to express a CAR on their surface, targeting tumor-specific antigens of cancer cells. The modified CAR-T cells are amplified in vitro and then infused back into the patients ( Figure 1 ) ( 21 ). Subsequently, CARs can identify and bind to specific antigens expressed on cancer cells and consequently eliminate and kill cancer cells ( 22 , 23 ).
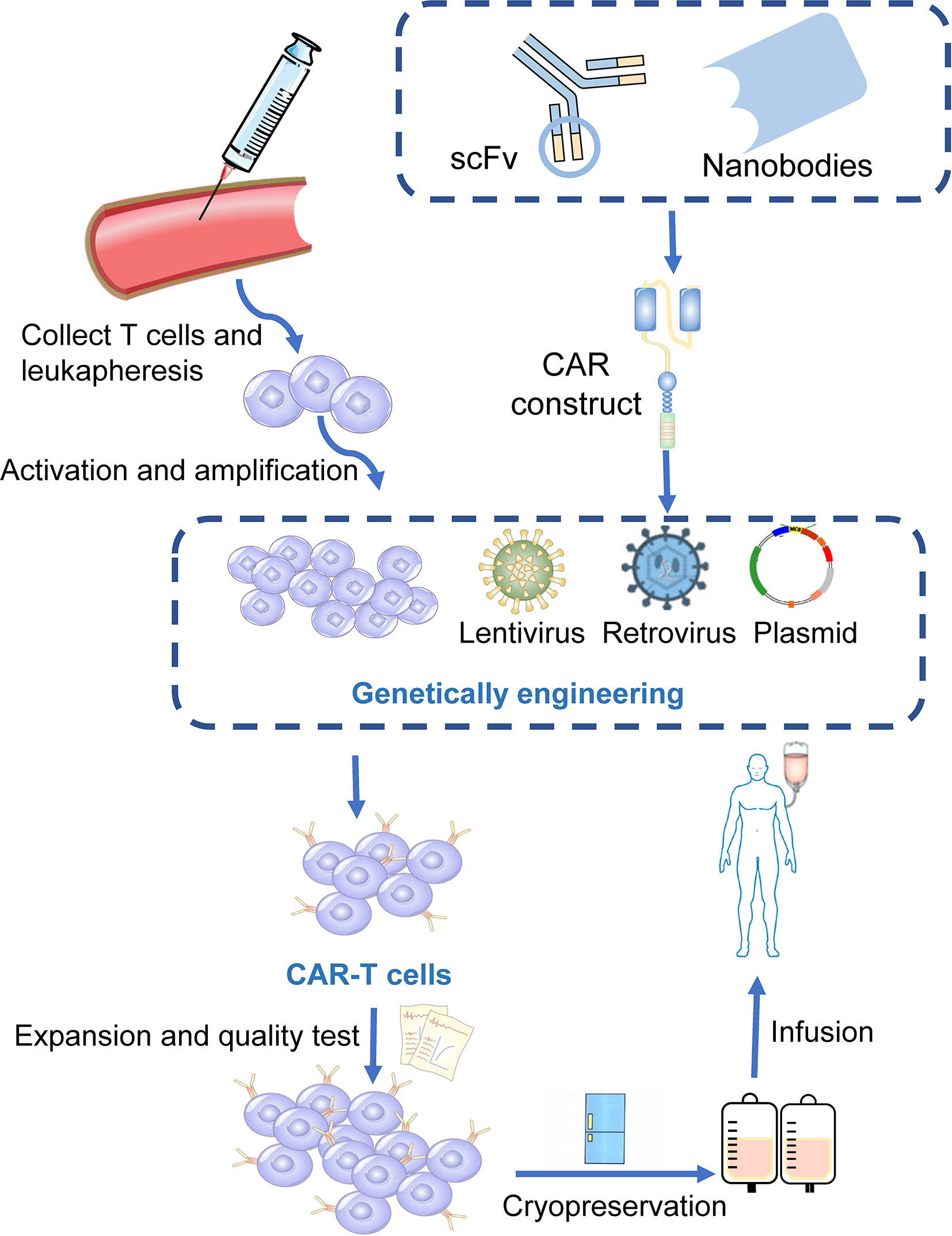
Figure 1 Manufacturing procedures of CAR-T cells. T cells are firstly collected from the peripheral blood of the patients. The activated and amplified T cells are genetically engineered with CAR structure via retroviral, lentivirus or other vectors. CAR-T cells are then expanded ex vivo and a quality control procedure is applied. Finally, those modified T cells were infused back into the patients.
CAR-T cell therapy is an emerging method against hematological malignancies and has demonstrated satisfactory curative effects, which is a substantial breakthrough in adoptive cell therapy ( 24 , 25 ). CAR-T cells targeting CD19 have become a leading engineered T-cell therapy strategy against relapsed or refractory acute lymphocytic leukemia and B-cell non-Hodgkin lymphoma ( 26 , 27 ). Yescarta (axicabtagene ciloleucel) and Kymriah (tisagenlecleucel) are currently approved to treat B-cell-derived malignancies, with response rates greater than 80% ( 28 , 29 ). Recently, Tecartus (brexucabtagene autoleucel) has also been approved for the treatment of adult mantle cell lymphoma ( 30 , 31 ). However, only targeting CD19 did not show considerable efficacy in most refractory multiple myeloma (MM) patients, partly due to the lower expression of CD19 on the cell surface of myeloma, and there is no FDA-approved CAR-T cell therapy against it ( 22 , 32 , 33 ). Clinical trials have indicated that CD269 (B cell maturation antigen, BCMA) and CD138 (also known as syndecan 1) molecules, which are mostly expressed in mature B cells or plasma cell surfaces, could exert substantial anti-MM activity ( 34 – 36 ). The unprecedented achievements of CAR-T cell therapy in hematological malignancies have also improved the use of CAR-T cells in various solid tumors.
1.1 The Design and Development of CAR Structure
CARs are artificial fusion proteins that comprise four major parts: extracellular antigen recognition and binding domains, spacer/hinge domains, transmembrane domains, and intracellular signaling domains ( 37 , 38 ). Every component of the CAR structure has unique properties and has evolved to optimize the CAR function ( 39 ). The extracellular domains are responsible for recognizing and binding the targeted tumor-specific antigens, whereas intracellular signal domains primarily induce T-cell proliferation and corresponding signal transduction ( Figure 2 ) ( 40 ). Recently, armored CAR-T cells have been engineered to overcome immunosuppressive tumor microenvironment (TME) ( 41 ). Engineered CAR-T cells can secrete various cytokines such as IL-12, chemokines, or co-expressing immunomodulatory ligands to alter the inhibitory microenvironment in the TME and support CAR-T cell function ( 20 ).
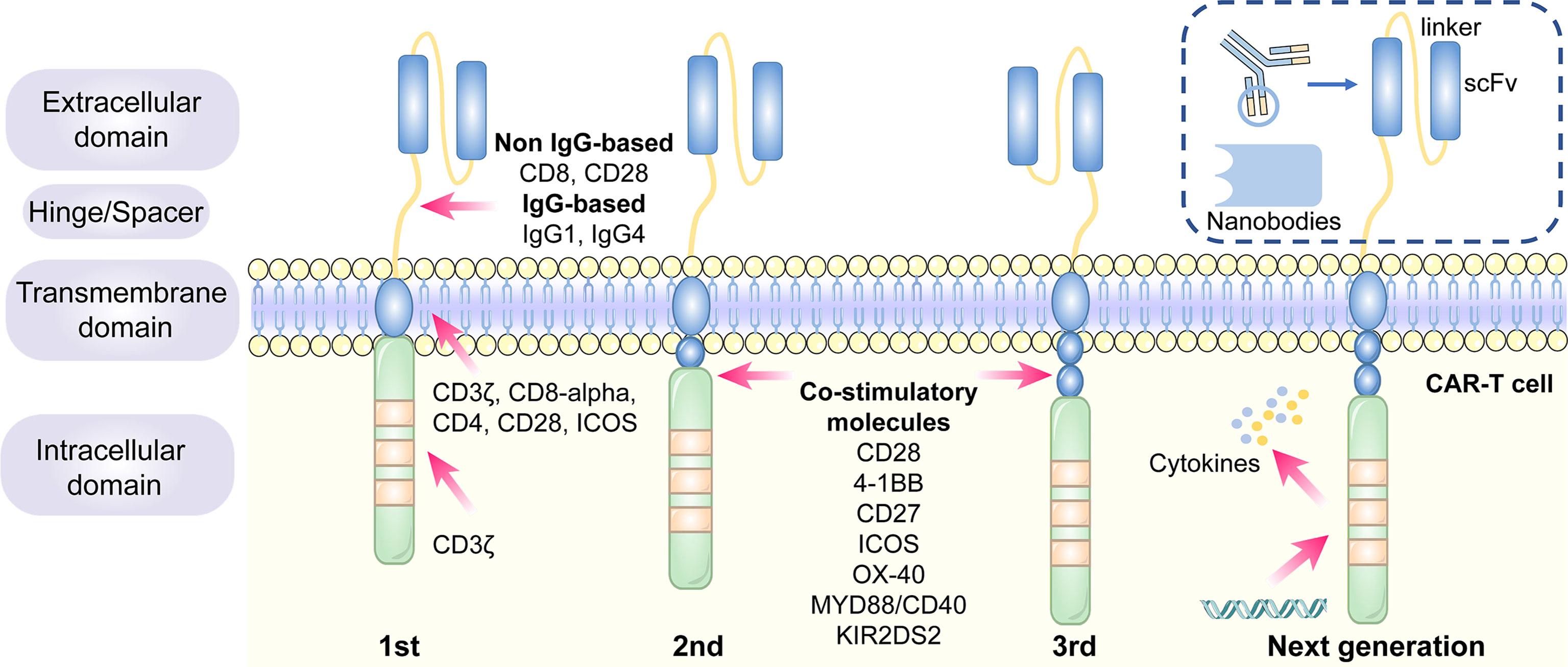
Figure 2 The structure and evolution of CAR-T cells from the first generation to the fourth generation. The CAR-T cells are consisted of extracellular tumor antigen binding domains (scFv, nanobodies), hinge regions, transmembrane regions and intracellular signaling domains. Different generations of CAR structures are primarily characterized by distinct intracellular signaling domains. The first generation of CAR-T cells only contain a CD3ζ intracellular signaling domain, with less persistence and efficacy in clinical practice. The second or third generation of CAR-T cells include one or more costimulatory molecules, and the next generation of CAR-T cells are engineered to express cytokines, which greatly improve their competence to eliminate the tumor cells.
1.1.1 Antigen Recognition and Binding Domains
The single-chain variable fragment (scFv) is derived from the variable heavy and variable light chains of a monoclonal antibody connected by a flexible linker ( 42 ). It is the major component of the extracellular antigen recognition and binding moieties, which can effectively recognize tumor antigen targets in a major histocompatibility complex (MHC)-independent manner and trigger CAR downstream signaling and CAR-T cells ( 43 ). The scFv sequences determine the specificity and binding affinity of the targeted antigens of the CAR ( 44 ). The high affinity of scFv has been reported to result in on-target, off-tumor toxicity, and severe cytokine release syndrome ( 45 ). Moreover, scFv can be designed to bind to soluble ligands, such as transforming growth factor-beta (TGF-β), contributing to the conversion of the immunosuppressive role of TGF-β ( 46 ). Single-domain antibodies (known as nanobodies or VHHs), whose variable regions only contain heavy chains instead of light chains, are stable camelid-derived single-domain antibodies ( 47 ). They are smaller in size and have a similar affinity to traditional scFv; however, they avoid the shortcomings of traditional scFv, such as low folding efficiency and tendency to aggregate ( 48 , 49 ). In addition, cytokines ( 50 ), ligands ( 51 – 54 ) and antigen recognition peptides (adnectins and designed ankyrin repeat proteins) could be applied as an option for antigen recognition and binding regions of CARs ( 55 , 56 ).
1.1.2 Hinge Domains
The length of the hinge regions can be adjusted to optimize the distance between CAR-T cells and targeted tumor cells, ensuring the folding efficiency of CAR scFv and providing a flexible and persistent connection for CAR signal transduction ( 57 ). In addition, the domains also augment the binding affinity of CAR-T cells and targeted cells ( 38 ). Hinge domains play a crucial role in regulating the expression and transport efficiency of CAR and the definition of the CAR signaling threshold ( 57 ). The spacer domains enable the CAR to access target epitopes that are otherwise sterically inaccessible ( 58 ). They can also be used to modulate synaptic cleft distances, as distal membrane antigen epitopes commonly require shorter spacers, whereas proximal membrane antigen epitopes require longer spacers ( 58 , 59 ). Non-IgG-based spacers, including CD8 and CD28, and IgG-based spacers, such as IgG1 or IgG4, have been proven to be equally effective and are utilized in the construction of CAR hinge domains ( 58 , 60 ). The spacers containing Fc domains must be changed after recognizing the targeted antigens, in case of in vivo interactions with cells expressing Fc gamma receptors that result in off-target activation of CAR-modified T cells or impaired antitumor efficacy ( 61 ).
1.1.3 Transmembrane Domains
The transmembrane domains serve as anchors to connect the extracellular antigen-binding domain to the cell membrane and transduce extracellular antigen-recognition signals to the intracellular domains ( 38 , 58 ). They primarily originate from type I transmembrane proteins, including CD3ζ, CD8-alpha, CD4, or CD28 ( 20 , 62 ). The stability and function of CARs are associated with transmembrane domains ( 38 ). Bridgeman et al. reported that CARs containing the CD3ζ transmembrane domain can form a complex with endogenous T cell receptor (TCR), and subsequently, may induce T cell activation ( 63 ). In vivo studies indicated that CD8-alpha resulted in lower levels of inflammatory cytokines and T-cell activation-induced death than CD28 ( 64 ). CD28 is currently the most stable transmembrane domain ( 39 ). Third-generation CAR T cells carry a B7-family inducible costimulator (ICOS) transmembrane domain ( 65 ). The persistence and anti-tumor activity of CAR-T cells is substantially promoted when the ICOS transmembrane domain is connected to an ICOS intracellular domain ( 62 ).
1.1.4 Intracellular Signaling Domains
The endodomains normally comprise a CD3ζ transducer, and one or more co-stimulatory signaling molecules such as CD28, 4-1BB (CD137), CD27, ICOS, OX-40, MYD88/CD40, and KIR2DS2 ( 66 ). This design pattern further prolongs the survival time and promotes the proliferation and antitumor activities of CAR-T cells ( 38 , 67 , 68 ). CD28 and 4-1BB, fused to the intracellular CD3ζ domain, are the most extensively studied and intensively applied co-stimulatory molecules ( 69 ). However, their clinical efficacy is far from each other. CAR-T cell therapy based on 4-1BB costimulatory domain is generally admitted to have more superior clinical efficacy, because 4-1BB costimulatory domain could ameliorate the exhaustion mediated by CAR signaling ( 70 , 71 ). CAR-T cell product based on CD28 costimulatory domain initiates faster antitumor property, while compared with 4–1BB costimulatory domain, it is less persistent since fewer central memory T cells are formed ( 72 ) ( Table 1 ). Additionally, CAR-T cells, incorporated two costimulatory molecules, such as ICOS and 4-1BB, have showed tremendous efficacy in preclinical mouse models ( 62 , 73 ).The other co-stimulatory signaling molecules, including CD27 ( 74 , 75 ), OX-40 ( 76 , 77 ), MYD88/CD40 ( 78 ) and KIR2DS2 ( 79 ) have demonstrated promising efficacy in preclinical models but have not been tested in clinical trials.

Table 1 Comparison of properties of different costimulation 4-1BB versus CD28 in CAR-T cell.
1.2 The Generation of CAR−T Cells
Different generations of CAR structures, characterized by distinct intracellular signaling domains, have been designed to improve the safety and efficacy of CAR-T cell therapy against various cancers ( 80 ). First-generation CAR-T cells only contain one intracellular signaling domain, CD3ζ, with less impressive clinical efficacy for the lack of persistence and proliferative activity ( 38 ). Inclusion of the costimulatory molecules equipped with second-generation CAR-T cells with the necessary signals for activation considerably prolonged the survival time of CAR-T cells and improved clinical outcomes in cancer patients ( 81 ). Third-generation CAR-T cells aggrandize a costimulatory molecule compared with second-generation CAR-T cells, consisting of CD3ζ and two costimulatory molecules (CD27, CD28, 41BB, ICOS, OX-40, etc.), further augmenting and enhancing their competence to clear tumor cells ( 82 , 83 ). In particular, the fourth generation of CAR-T cells known as T cells redirected for universal cytokine-mediated killing (TRUCK), which can recruit nuclear factor of activated T cells (NFAT) to induce the release of cytokines IL-12 IL-15 and granulocyte–macrophage colony-stimulating factor ( 84 ). The anti-tumor activity of the fourth generation of CAR-T cells is enhanced by overcoming the immunosuppressive effect of the TME ( Figure 2 ) . The fifth-generation CAR-T cells, which is proposed to remove the TCR alpha and beta chains through gene editing technology, avert the risk of graft-vs.-host disease, and manufacture “off the shelf” products, are still under investigation ( 85 ).
Although the structure of CARs is constantly evolving to promote efficacy and diminish the cytotoxic effects of CAR-T cell therapy, second-generation CAR-T cells still remain the mainstay of clinical application ( 86 ).
1.3 NSCLC and SCLC−Associated Antigens for CAR−T Cell Therapy in Preclinical Studies
CAR-T cell therapy has emerged as a novel approach to adoptive cell immunotherapy in recent decades. In solid cancers, it is more complex to construct CAR-T cells because it is difficult to identify tumor-specific antigens to be targeted. Several surface antigens have already been evaluated in preclinical studies as potential CAR-T cell therapy targets. Thereafter, we provide detailed descriptions of several novel targets.
1.3.1 Mesothelin (MSLN)
MSLN, a tumor differentiation antigen with the low expression on normal mesothelial cells, is overexpressed in a wide range of solid cancers, including lung cancer, mesothelioma, and pancreatic carcinoma; therefore, it could be used as a potential target ( 87 , 88 ). High expression of MSLN is commonly correlated with negative clinical outcomes in NSCLC ( 67 ). In ex vivo experiments, MSLN-targeted CAR-T cells exerted substantial inhibitory effects on cancer cell proliferation and invasion ( 89 ). The efficiency of MSLN-targeted CAR-T cell therapy has been assessed in subcutaneous mouse lung cancer models ( 90 ). A slower growth rate of tumor size was observed in the tail vein injection of MSLN-targeted CAR-T cells ( 89 ). In summary, MSLN-targeted CAR-T cells could be feasible for MSLN-positive cancers, such as NSCLC.
EGFR belongs to the HER/ErbB family of receptor tyrosine kinases that transduces extracellular growth signaling into the cells ( 91 ). More than 60% of NSCLC patients harbor activating EGFR mutations, contributing to the overexpression of EGFR, making it possible to target EGFR as a treatment for CAR-T cell therapy against NSCLC ( 91 ). EGFR-CAR T cells were found to exhibit greater cytotoxic activity in vitro ( 92 ). In nude mouse subcutaneous xenografts, EGFR-CAR T cells dramatically decreased tumor size and volume ( 93 ). The above results indicate that EGFR-targeted CAR-T cell therapy could be applied to NSCLC patients in the future ( 94 ).
1.3.3 Receptor Tyrosine Kinase-Like Orphan Receptor 1 (ROR1)
ROR1 is a crucial oncofetal glycoprotein that can sustain pro-survival and pro-apoptotic signaling in lung adenocarcinomas ( 95 , 96 ). It has been proposed as a targeted antigen in CAR-T cell therapy as the overexpression of ROR1 protein has been observed in various malignancies, including lung cancer ( 97 , 98 ). ROR1-CAR T cells maintained their anti-tumor activity, cytokine secretion, and proliferation in NSCLC models in vitro and in vivo ( 97 , 99 ). Carolina et al. demonstrated the safety and function of second-generation ROR1 CAR-T cells in macaques ( 100 ).
1.3.4 Mucin-1 (MUC1) and Prostate Stem Cell Antigen (PSCA)
Aberrant high expression of MUC1 regulates the expression of programmed death-ligand 1 (PD-L1) in cancer cells, which could prevent cancer cells from being cleared by the immune system ( 101 , 102 ). PSCA, a glycosylphosphatidylinositol (GPI)-anchored cell surface protein, belongs to the Thy-1/Ly-6 family ( 103 ). MUC-CAR T cells and PSCA-CAR T cells identify and eliminate PSCA+ or MUC1+ NSCLC cells, respectively, in vitro ( 104 ). PDX mouse subcutaneous models generated from NSCLC patients whose tumors only express PSCA or both PSCA and MUC1 were applied to explore the efficacy of PSCA and MUC1 CAR-T cells against NSCLC. Tumor growth was substantially inhibited in CAR-PSCA T cells. Thereafter, a combination of PSCA and MUC1 CAR-T cells exerted a synergistic effect on tumor survival ( 104 ). Therefore, MUC1 and PSCA could be promising CAR-T cell therapy targets for the treatment of NSCLC.
1.3.5 Human Epidermal Growth Factor Receptor 2 (HER2)
HER2 belongs to the HER/ErbB family of receptor tyrosine kinases involved in cell proliferation and angiogenesis ( 105 ). The anti-tumor effect of HER2 CAR-T cells against two NSCLC cell lines, A549 and H1650, was observed in a 96-h co-culture assay ( 106 ). Moreover, in orthotopic or subcutaneous A549 NSCLC mouse xenograft models, HER2 CAR-T cell therapy decreased tumor growth and could not completely eliminate tumors ( 106 , 107 ).
1.3.6 Carcinoembryonic Antigen (CEA)
CEA is an oncofetal glycoprotein generally expressed during fetal development; however, its expression declines after birth ( 108 ). CEA levels increase rapidly in the tumorigenesis and development of lung cancer ( 109 ). Therefore, preclinical studies of CAR-T cell therapy targeting CEA have been conducted. CEA-targeted CAR-T cells have been found to eradicate advanced lung carcinomas ( 110 ).
1.3.7 PD-L1
Immunotherapy targeting programmed death-1(PD-1)/PD-L1 signaling has achieved substantial progress in NSCLC treatment. Accumulating evidence shows that PD-L1, both in tumor cells and in the TME, suppresses T cell proliferation and mediates anti-tumor immunity ( 111 ). PD-L1-targeted CAR-T cells exhibited robust cytotoxic effects against NSCLC cells in vitro and in vivo ( 112 , 113 ). Therefore, PD-L1-targeted CAR-T cells could be a novel curative approach for PD-L1-positive NSCLC patients.
1.3.8 Fibroblast Activation Protein (FAP)
FAP is a marker expressed on cancer- associated fibroblasts (CAFs) in a majority of human malignancies ( 114 ). FAP molecule itself and FAP-positive cells in TME could contribute to cancer cell proliferation, invasion, angiogenesis and extracellular matrix (ECM) remodeling ( 115 ).
FAP targeted CAR-T cells inhibited the proliferation of TC1 and A549 lung cancer cells by eliminating FAP-positive stromal cells in mice models ( 114 , 116 ). In contrast, another study claimed that FAP targeted CAR-T cell achieved limited antitumor efficacy and severe side effects for bone marrow stromal cells (BMSCs) were also being killed ( 117 ). Therefore, the feasibility of targeting FAP as a specific antigen in CAR-T therapy remains to be verified.
1.3.9 Other Targeted Antigens
Several tumor antigens, such as lung-specific X (LUNX), variant domain 6 of CD44 gene, melanoma-associated antigen-A1 (MAGE-A1), erythropoietin-producing hepatocellular carcinoma A2 (EphA2), and glypican-3 (GPC3), are under active investigation for application as targeted antigens of CAR-T cell therapy against NSCLC ( 118 – 122 ). For SCLC, CD56-and Delta-like ligand 3 (DLL-3)-targeted CAR-T cells are being explored ( 123 , 124 ). Bivalent tandem CAR-T cells are equipped with two targeted antigens. CD70, B7-H3, MUC1, PSCA, PD-L1, and CD80/CD86, have exhibited enhanced antitumor efficacy in lung cancer ( 104 , 125 ). B7-H3 is one of inhibitory ligands, which belongs to B7 immunoglobulin family. Although its corresponding immune checkpoint receptors remain undetermined, the inhibitory role of B7-H3 has been confirmed in preclinical studies ( 126 ). The expression of B7-H3 is aberrantly augmented in a wide range of solid tumor tissues, compared with normal tissues, which supports the possibility of targeting B7-H3 in CAR-T cell therapy against lung cancer ( 125 , 127 ). CD80/CD86 are immune checkpoint ligands shared by inhibitory CTLA-4 and costimulatory CD28. CD80/CD86-targeted CAR-T cells have been generated to reverse the inhibitory CTLA4-CD86/CD86 signals and prevent the survival of B cell malignancies and other tumors including NSCLC ( 128 ). The efficacy of CAR-T cell therapy, which targets both tumor cells and tumor-associated macrophages in the TME, has also been validated in NSCLC ( 129 ) ( Figure 3 ).
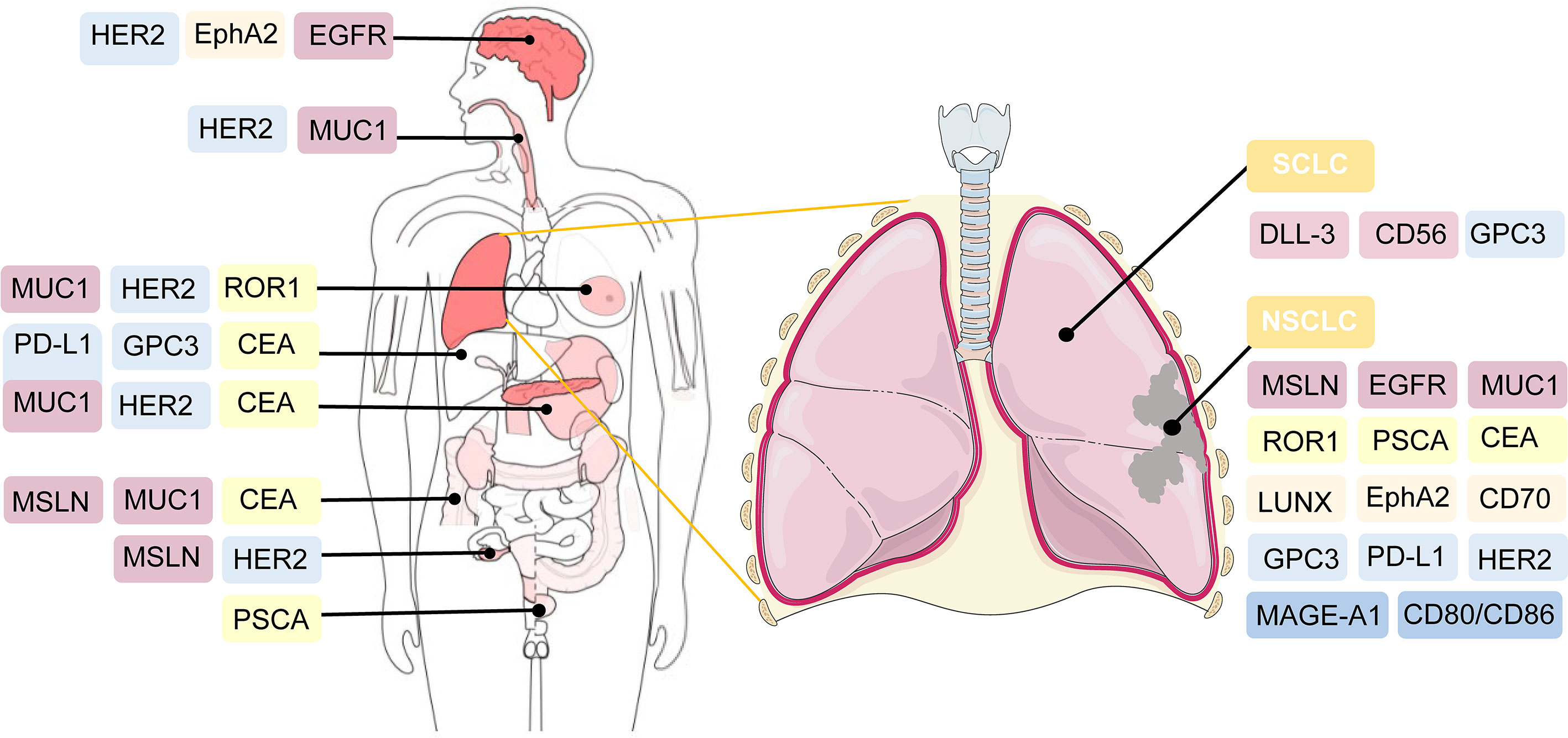
Figure 3 Potential targeted antigens for CAR-T cell therapy in preclinical and clinical trials. In the right, antigen targets are listed against SCLC and NSCLC. As shown in the left of the figure, these antigens are also broadly applied in CAR-T cell therapy against other solid tumors.
1.4 NSCLC and SCLC−Associated Antigens for CAR−T Cell Therapy in Clinical Trials
CAR-T cell treatment has achieved substantial success against several hematological malignancies. At present, the primary task is to broaden the applications of CAR-T cell therapy from merely hematologic tumors to multiple solid tumors. Thus, its safety and efficacy in solid cancers are under intensive investigation. The feasibility of CAR-T therapy against solid tumors is currently being evaluated in approximately one-third of CAR-T clinical trials ( 130 ). Among them, the majority are on CAR-T therapy for the treatment of lung cancer. The extraordinary progress of CAR-T therapy for lung cancer is promising; however, many challenges and hurdles exist. Therefore, the clinical application of CAR-T in NSCLC and SCLC treatment is still under intensive exploration. The optimal target for CAR-T cell therapy is specifically expressed or generally overexpressed in tumor cells, whereas it is expressed at very low or limited levels in normal peripheral cells or tissues ( 131 ). Current clinical trials of CAR-T therapy against NSCLC and SCLC primarily focus on MSLN, MUC1, GPC3, PSCA, EGFR, CEA, HER2, PD-L1, ROR1, and other promising targets ( Table 2 ) .
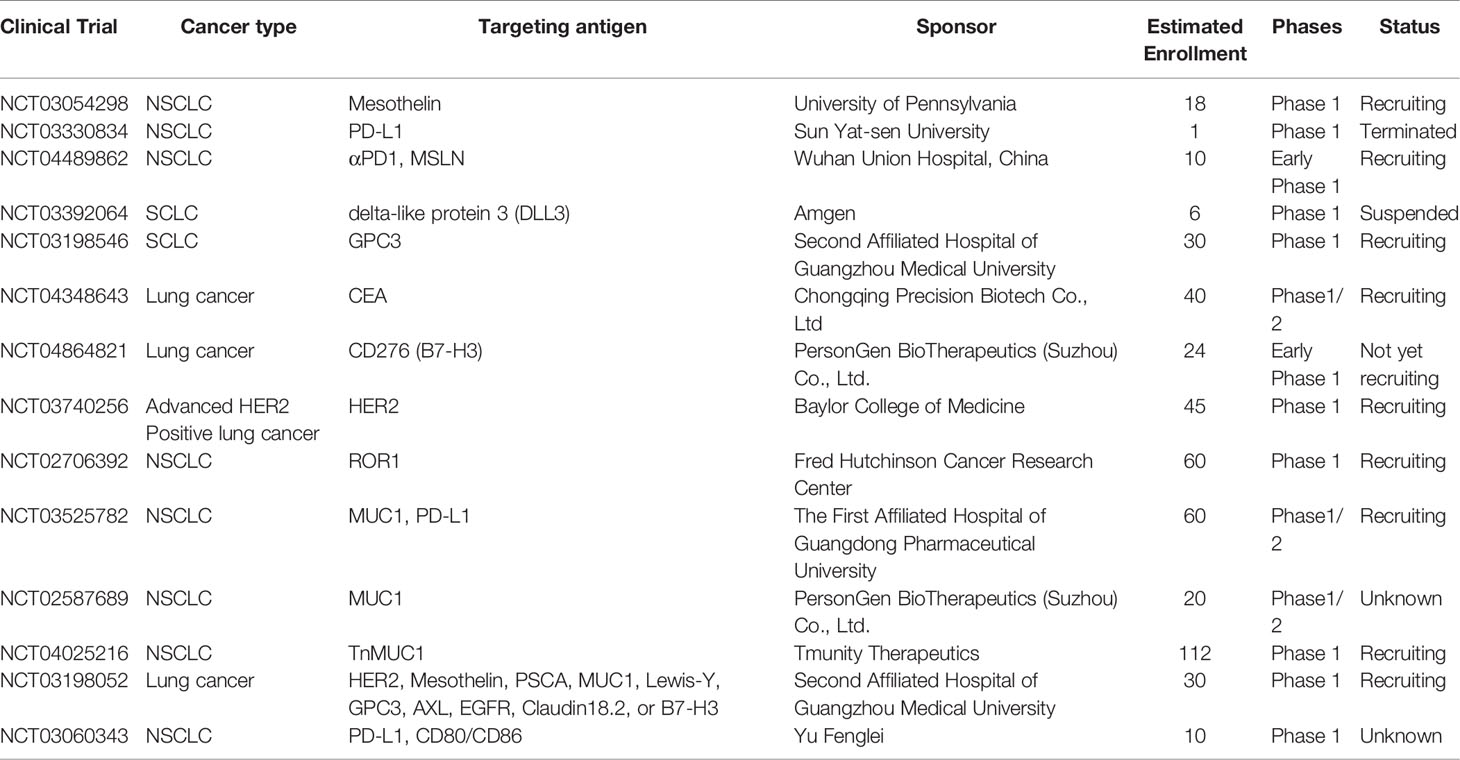
Table 2 Underlying targeting antigens of NSCLC and SCLC for CAR-T cell therapy in clinical trials.
2 Challenges and Engineering Strategies
Over the past few years, there has been a rapid increase in the use of CAR-T cell therapy to treat hematological malignancies and solid tumors. Many clinical trials have made substantial achievements; however, severe therapeutic responses to CAR-T cell therapy and unsatisfactory treatment efficacy hinder rapid development. In 2010, a patient with multiple metastases of colon cancer died after administering CAR-T cells targeting ERBB2. The patient experienced respiratory distress within 15 min after CAR-T cell transfusion and died five days after the treatment ( 132 ). Compared with hematological malignancies, solid tumors face a unique set of challenges, including issues confusing hematological malignancies, more severe and complicated related toxicities, the lack of a strongly expressed tumor-associated antigen target, low infiltration of T cells in tumor tissue, CAR-T cell exhaustion, and a highly immunosuppressive and metabolically challenging TME, which limit the safety and effectiveness of treatment ( 133 – 135 ). Future studies to develop practical engineering strategies to enhance the efficacy of CAR-T cell therapy and minimize adverse reactions should be conducted.
2.1 Overcoming Treatment-Related Toxicities
CAR-T cell therapy can result in a range of toxicity events. The major treatment-related toxicities include cytokine release syndrome (CRS) and immune effector cell-associated neurotoxicity (ICANS), which particularly peak in the first or second week of CAR-T cell administration, respectively ( 133 ). Patients with CRS mostly have common manifestations such as fever, tachycardia, hypoxia, dyspnea, hypertension, coagulopathy, and elevated serum cytokines, including interleukin-6 (IL-6) ( 136 , 137 ). ICANS is characterized by tremor, encephalopathy, cerebellar alteration, or seizures ( 138 ). Both CRS and ICANS are caused by the activation of CAR-T cells and cytokines secreted by the associated immune cells. CAR-T cells can release pro-inflammatory cytokines, including IL-2, IL-6, and IFN-γ, and then activate more immune cells to secrete IL−1RA, IL−10, IL−6, IL−8, IFNα, and other cytokines, which eventually could lead to massive cytokine release ( 139 ). Hemophagocytic lymphohistiocytosis/macrophage activation syndrome has also been reported following CAR-T cell therapy. It is characterized by hyperinflammatory syndrome and multiple organ dysfunction ( 140 ). IL−6/IL−6R antagonists and corticosteroid usage can interrupt the inflammatory process and play a substantial role in symptom remission ( 141 ). It is critical to detect these treatment-related toxicities early and provide appropriate treatment based on the toxicity grade as soon as possible.
Selecting co-stimulatory signaling molecules and transmembrane domains could have an impact on cytokine production and CAR-T cell function. Compared with CD28/CD3ζ CAR T cells, 4–1BB/CD3ζ CAR T cells amplified more slowly, persisted for a longer time, and secreted less cytokines ( 142 ). CAR-T cells with CD8-alpha transmembrane domains have been shown to release less cytokines than those with CD28 domains ( 64 ). In addition, the inclusion of inducible caspase-9 safety switches to CARs has been verified to control the expansion of CAR-T cells and the load of cytokines ( 143 ). In summary, genetic modification of CAR designs might help reduce the generation of cytokines and the incidence of treatment-related toxicities ( Figure 4 ).
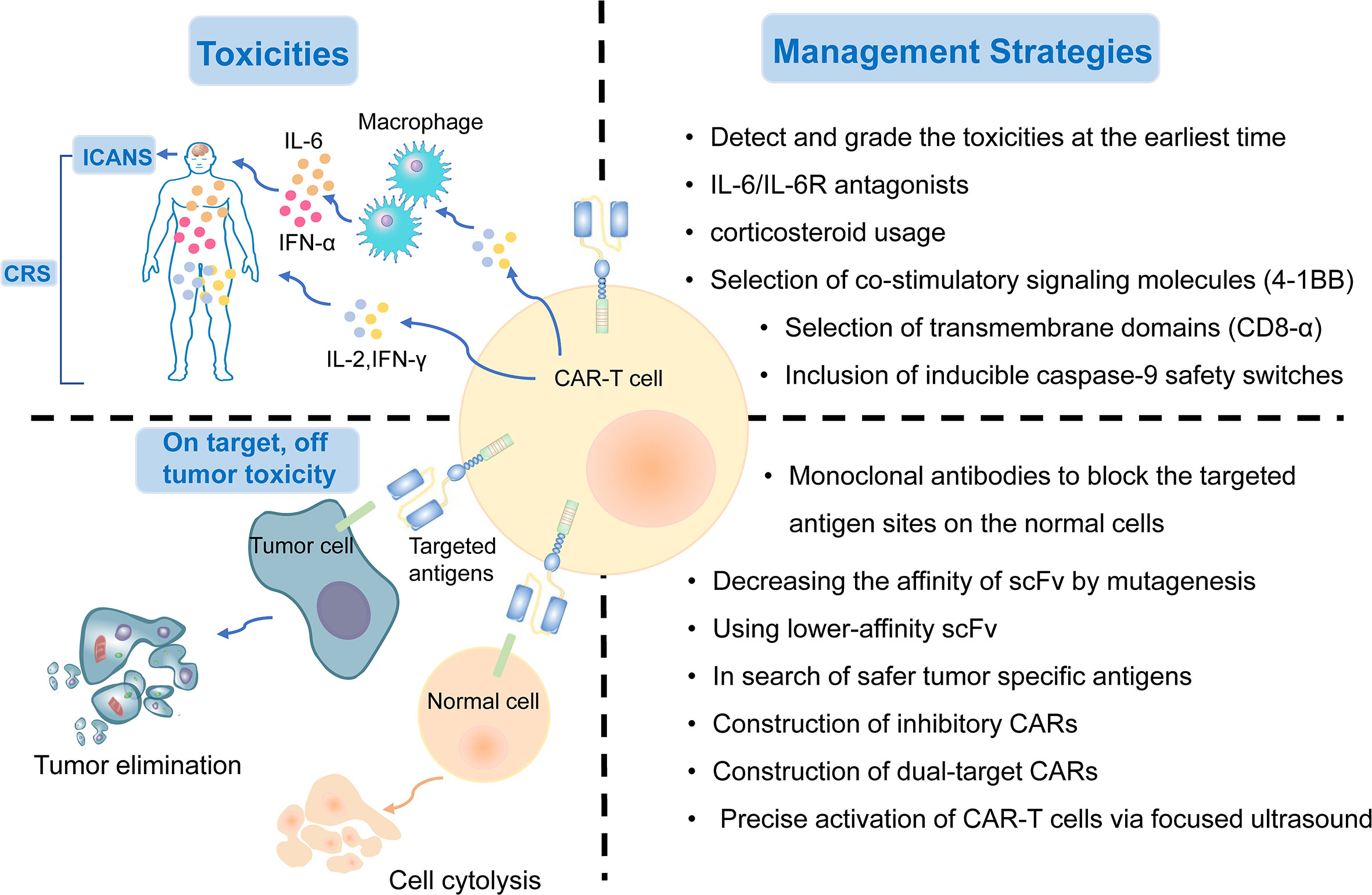
Figure 4 The treatment-related and on-target, off-tumor toxicities and corresponding management strategies of CAR-T cell therapy.
2.2 On-Target, Off-Tumor Toxicity
Although the targeted tumor-associated antigens are carefully screened, many normal cells still suffer from the attack of T cells because they express the same or similar antigens. On-target, off-tumor toxicity, manifesting multiple organ injury and failure, is an issue impeding the development of CAR-T cell treatment. Thus, there is an urgent need to explore safer targeted tumor-associated antigens for lung cancer treatment. To date, MSLN, EGFR, ROR1, MUC1, PSCA, and HER2, as described previously, are the most targeted antigens in CAR-T cell therapy for NSCLC. Several other tumor antigens, including LUNX and B7-H3, also exhibit great potential as targeted antigens in CAR-T cell therapy because they are aberrantly expressed in lung cancer tissues, with a relatively low expression in normal tissues ( 118 , 144 ).
The on-target toxicity is antigen-oriented, and shielding of a CAR-targeted antigen expressed on normal tissues could minimize toxicity and optimize the efficacy of CAR-T cell therapy. Some renal cell carcinoma patients developed hepatic enzyme disorders that required discontinuation of therapy after receiving anti-carbonic anhydrase IX (CAIX) CAR-T cell therapy. This on-target toxicity can be overcome by pre-administration of parental anti-CAIX monoclonal antibodies to block the CAIX antigen sites in the liver ( 145 ). In addition, decreasing the affinity of scFv by mutagenesis or using lower-affinity scFv as a replacement could also substantially reduce on-target, off-tumor reactivity without affecting the antitumor activity ( 45 ). Other attempts include the construction of inhibitory CARs, which could protect the normal cells from being attacked by targeted CAR-T cells, and dual-target CAR-T cells, which require two signals to be full activated ( 146 ). Recently, an inducible CAR-T cell, was developed to be activated via focused ultrasound within specific tumor sites, which could dramatically mitigate the on-target, off-tumor toxicity, in comparison to conventional CAR-T cells ( 147 ) ( Figure 4 ).
2.3 Evasion of Antitumor Immune Responses
A common mechanism for tumor cells to evade immune surveillance in CAR-T cell therapy is the downregulation or even loss of targeted antigens, whose expression level could exert a direct impact on the therapeutic efficacy ( 148 ). Targeting CD19/CD20 CAR-T cell therapies have led to promising achievements in treating B-cell malignancies in recent years ( 149 ). Tumor-associated antigens in hematologic malignancies are highly expressed and easier to target, whereas antigens in solid tumors have greater heterogeneity and lower expression levels, making it difficult to eliminate solid tumor cells ( 150 ). Intratumor heterogeneity might be a key factor contributing to the evasion of antitumor immune responses ( 151 ). In lung cancer, common targets such as MSLN, MUC1, PSCA, and epithelial cell adhesion molecule, have intratumoral heterogeneity, leading to an unsatisfactory outcome of CAT-T cell therapy in lung cancer ( 21 ). Many clinical studies have shown that when tumors relapse after treatment, tumors are found to undergo antigen loss or become antigen-negative ( 50 , 152 ). This phenomenon may be mediated by the selective pressure applied by CAR-T cells to tumor cells, leading to the progressive selection of antigen-negative cells ( 82 ).
To overcome the evasion of antitumor immune responses, one approach is to engineer CARs with dual-specificity (i.e., simultaneously targeting two antigens) ( 153 ). Bispecific T cell-engagers (BiTEs), consisted of two scFvs, are produced by genetically engineered CAR-T cells to redirect both T cells and CAR-T cells against specific tumor cells ( 154 , 155 ). EGFRvIII-specific CAR-T cells secreting BiTE have shown to circumvent antigen escape in glioblastoma, and its effect on lung cancers remains to be further investigated ( 154 ). Tandem CAR-T cells can mitigate antigen escape and translate into superior antitumor activity ( 156 , 157 ) ( Figure 5 ). Armored CAR-T cells secreting pro-inflammatory cytokines, such as IL-18, have also been shown to elicit an enhanced antitumor immune response in preclinical models ( 158 ).
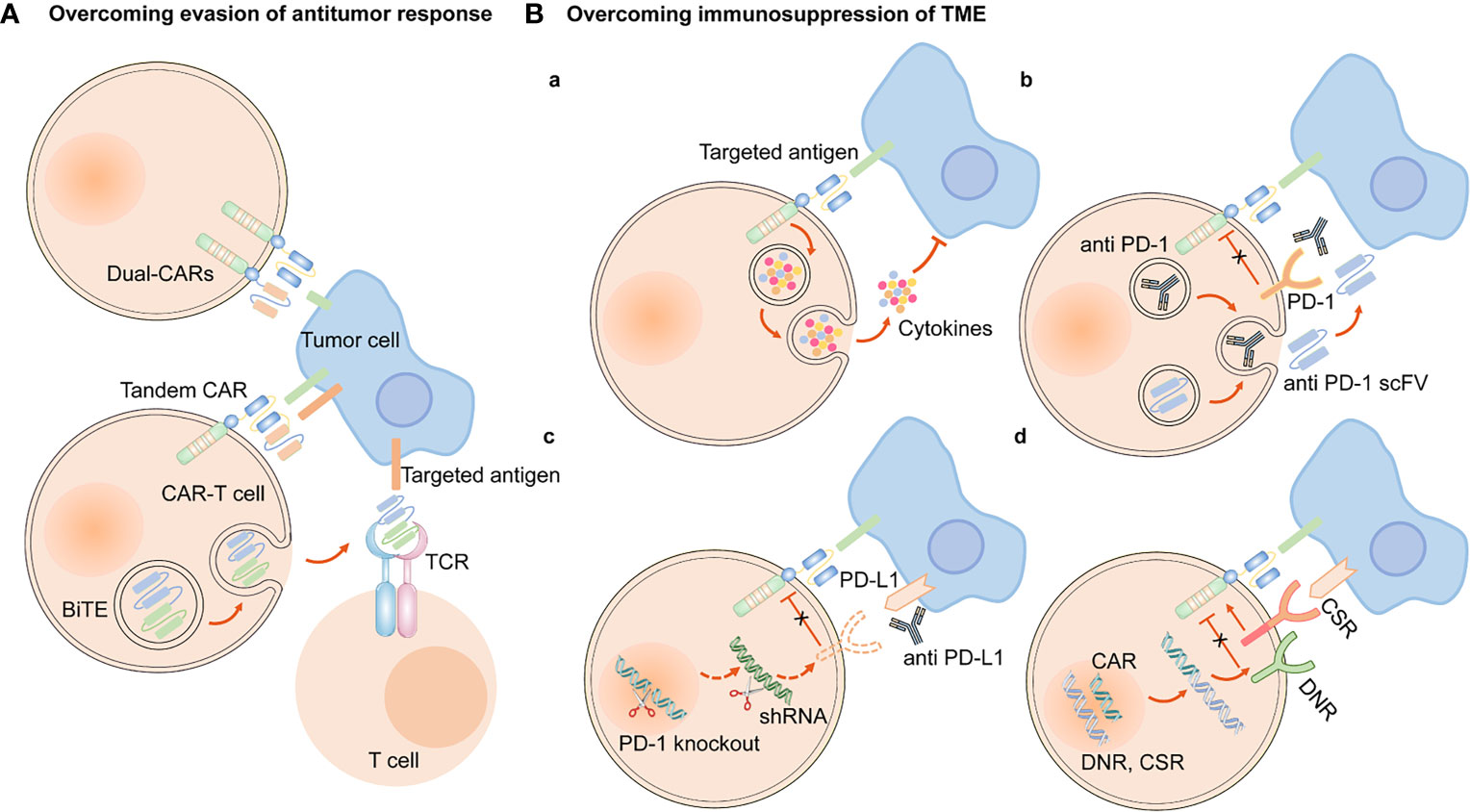
Figure 5 Engineering strategies to overcome evasion of antitumor response and immunosuppression of TME. (A) CAR-T cells are engineered to simultaneously target two antigens (dual CAR-T cells), and secret BiTE to redirect both T cells and CAR-T cells against specific tumor cells and circumvent antigen escape. Tandem CAR-T cells have bispecific receptors, which could target two different antigens. (B) (a) Armored CAR-T cells expressed immunostimulatory cytokines. Approaches to overcoming the immunosuppression of immune checkpoints in TME are as follows, (b) CAR-anti-PD-1/PD-L1 antibodies or scFv, (c) PD-1 gene knockout or downregulation of PD-1 expression by shRNA, (d) express a PD-1 DNR or a PD-1 CSR.
2.4 Physical Barriers
Cancer-associated fibroblasts (CAFs) and fibrotic environment contribute to the formation of physical barrier, preventing the CAR-T cells from being trafficked into tumor sites. Less infiltration of CAR-T cells into tumor tissues is another reason why the efficacy of CAR-T cell therapy in NSCLC is not as ideal as that in hematological malignancies.
CAFs are the predominant component of stromal cells in the TME and cannot be cleared by apoptosis ( 159 ). Owing to the heterogeneity of CAFs, they could play a dual role in pro-tumorigenicity and anti-tumorigenicity ( 160 ). They could regulate the growth, invasion, and angiogenesis of tumor cells by reshaping the ECM and secreting soluble growth factors ( 160 ). Moreover, growth factors, cytokines and chemokines, including fibroblast growth factor (FGF), TGF-β, C-X-C motif chemokine ligand 12 (CXCL12), and IL-6, are also secreted by CAFs to mediate immunosuppressive responses ( 161 ). Hence, they can be applied as potential targets for anticancer treatment. However, many challenges still prevail in modulating CAFs as an ideal target for CAR-T cell therapy. As previously mentioned, FAP-targeted CAR-T cell therapy induced lethal adverse effects because CAR-T cells attacked FAP-positive BMSCs ( 117 ). In addition, CAFs have been shown to contribute to the development of therapeutic resistance because the ECM produced by CAFs could serve as a thick barrier to block the penetration of drugs ( 162 ). Accordingly, we hypothesized that the physical barrier formed by CAFs could also hinder the delivery of CAR-T cells into tumor tissues, thus diminishing the effectiveness and efficacy of CAR-T cell therapy ( Figure 6 ).
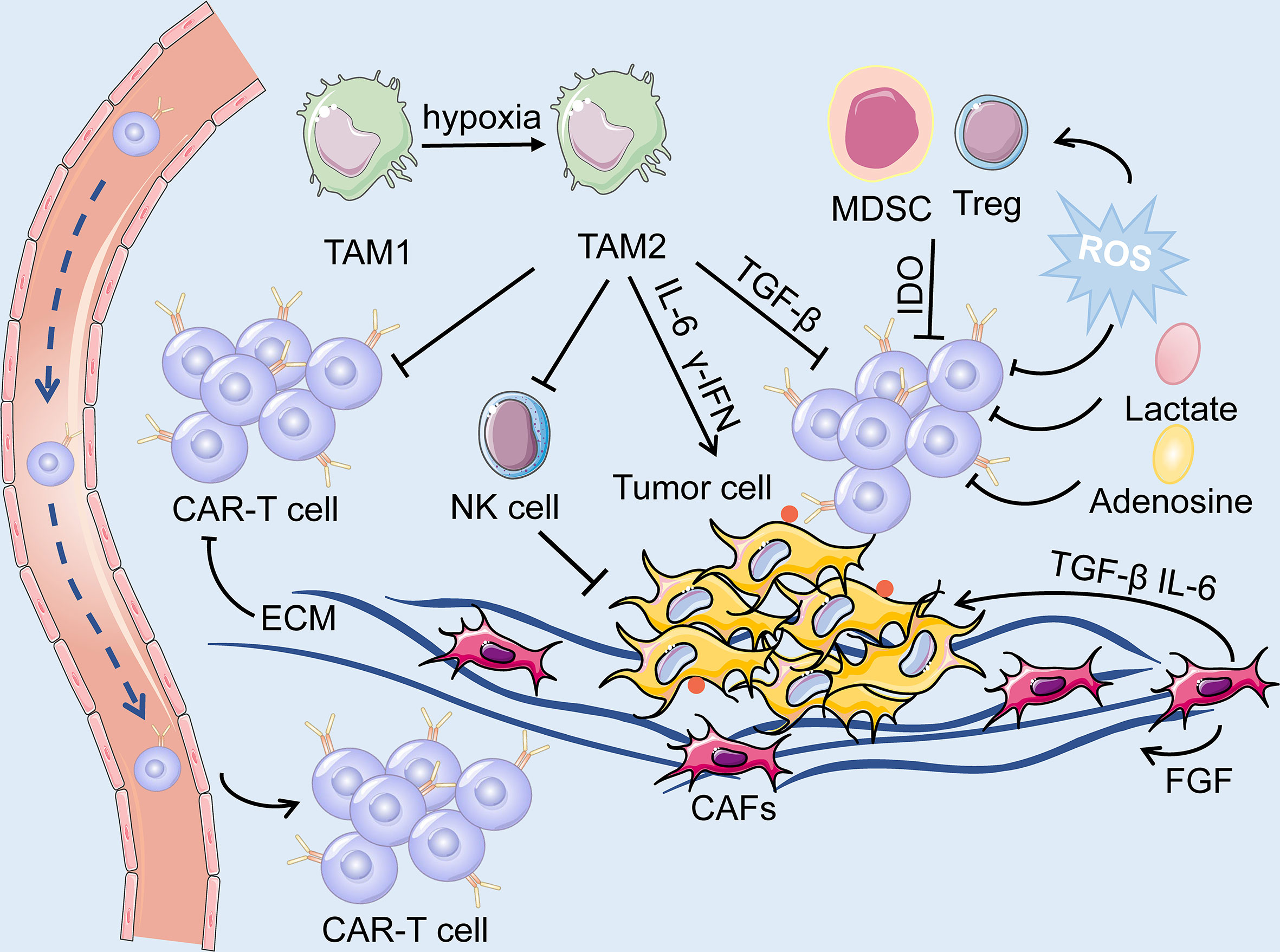
Figure 6 The TME is primarily composed of tumor cells, immune cells, immunosuppressive cells (including TAM, MDSC and Treg) and cytokines, CAFs, ECM and dysregulated tumor vasculatures. On the one hand, ECM produced by CAFs forms a physical barrier, impairing the infiltration of the CAR-T cells. On the other hand, the soluble cytokines secreted by the CAFs mediate immuno-suppressive responses, and consequently, facilitate the survival of tumor cells. The hypoxic and acidic environment directly deteriorate the metabolism of T cells while activating suppressive Tregs, leading to immunosuppression of CAR-T cells.
Several studies have been made to deplete or remodel the CAFs in the TME. One potential strategy is to apply FAP-redirected synthetic Notch CAR T cells or heparanase-modified CAR-T cells to deliver CAF remodeling molecules to suppress the expression profile of CAFs ( 163 ).
2.4.2 Fibrotic Environment
In contrast to hematological tumors, the infiltrative ability of CAR-T cells in lung cancer tissues is greatly restrained by the presence of a physical barrier. CAF activation, abnormal dense collagen, and ECM deposition contribute to developing a dense and fibrotic environment, altering the localization and migration of effector immune cells in NSCLC, which hinders immune cell infiltration and influences the efficacy of immunotherapy ( 164 , 165 ). In addition, the extensive fibrotic environment mostly lacks blood vessels, which creates a hypoxic TME and further impairs immune function ( 166 ).
The binding of chemokines and their corresponding receptors can mediate the trafficking of CAR-T cells through fibrotic environment. Hence, one approach to enhance the infiltration level of CAR-T cells is to engineer them to express chemokines or transgenic chemokine receptors ( 167 ). The CAR-T cells engineered to express IL-7 and CCL19 have been validated to increase the infiltration of peripheral CAR-T cells and dendritic cells and into tumor tissues and enhance the anti-tumor immune responses ( 168 ). Another engineering strategy is to construct enzyme-modified CAR-T cells to express heparanase, which accelerates the degradation of ECM and facilitates CAR-T cell trafficking to tumor sites ( 169 ). In addition, local injection of CAR-T cells is under investigation.
2.5 Immune Suppression in the TME
The TME of lung cancer has an immunosuppressive effect, as T cell activity is suppressed due to anti-inflammatory cytokines and upregulated immune checkpoint ligands. Additionally, the immunosuppressive cells, such as myeloid-derived suppressor cells (MDSCs), regulatory T cells (Tregs), tumor associated macrophages, and tumor associated neutrophils are broadly present in the TME ( Figure 6 ). CAR-T cell therapy against lung cancer is less efficient because of immune suppression of the TME and loss of CAR-T cell function.
One engineering approach to overcome the immunosuppressive role of TME is to establish armored CAR-T cells that secrete pro-inflammatory cytokines or chemokines, such as IL-12, IL-15, and IL-18 ( 20 ). These cells can recruit and activate innate immune cells such as natural killer (NK) cells and macrophages, and reprogram the immunosuppressive TME, which subsequently supports the proliferative and antitumor activity of CAR-T cells ( 170 ). In addition, based on blocking immune checkpoints, genetic knockdown of immune checkpoint receptors in CAR-T cells, such as PD-1, was demonstrated to enhance the anti-tumor effect. The clinical outcomes are being actively assessed in clinical trials on lung cancer ( 171 ). Other strategies include engineering CAR-T cells to secrete immune-checkpoint inhibitors, including anti–PD-1 scFv and anti–PD-L1 antibodies, to express PD-1 dominant-negative receptors (DNR) or PD-1 chimeric switch receptors (CSR) ( 113 , 172 , 173 ) ( Figure 5 ).
2.6 Metabolic Profile of the TME
Cumulating evidence supports that metabolism plays an essential role in the immune response because it could regulate the function and activity of T cells. The inhibition of T cell metabolism may directly deteriorate the activity of T cells while activating suppressive Tregs, resulting in immuno-suppression ( 174 ). The proliferation of CAR-T cells, secretion of cytokines, and elimination of tumor cells are all energy-demanding processes. However, tumor cells mostly consume a large proportion of energy and nutrients, while generate a mass of immunosuppressive metabolites, such as adenosine, lactate, and kynurenine ( 135 , 174 ). Moreover, indolamine-2,3-dioxygenase (IDO) secreted by tumor cells and MDSC could catalyze tryptophan into kynurenine, leading to the inactivation of CAR-T cells and the proliferation of Tregs ( 175 ) ( Figure 6 ). On the other hand, the dysregulated vasculatures also result in an extremely hypoxic and acidic TME. All of the above elements contribute to the formation of the metabolically hostile TME, which further impairs the function of CAR-T cells.
Reprogramming the CAR-T cells to adjust their metabolic properties through genetic or pharmacological inhibition of adenosine receptors A 1 and A 2A R substantially elevated CAR T cell efficacy in breast cancer, which appears to be a promising method to enhance CAR-T cell function in the TME ( 176 ). Additionally, ROS generated by MDSC exerts a negative impact on CAR-T cells, and therefore, the reduction of ROS might be a potential strategy to overcome the metabolic profile of TME. Furthermore, CD28 and 4-1BB, the co-stimulatory domains of CAR-T cells, respectively, improved the metabolic fitness of CAR-T cells in melanoma by upregulating the intake of glucose and the expression of glycolytic enzymes, and enhancing mitochondrial biogenesis and oxidative metabolism ( 177 , 178 ). However, limited data are available on the metabolic reprogramming of CAR-T cells in lung cancer.
2.7 CAR-T Cell Exhaustion
The existence of inhibitory ligands in the TME and endogenous TCRs leads to the gradual exhaustion of CAR-T cells ( 134 ). Clinical evidence has confirmed that CAR-T cell exhaustion markedly limits the efficacy of CAR-T cell therapy; therefore, it is imperative to prevent or reduce CAR-T cell exhaustion. However, it is difficult to reverse the cell exhaustion process directly by dedifferentiating T cells for exhaustion, which is a transcriptional and epigenetic forced differentiation state ( 179 ). Therefore, less differentiated T cell populations, such as naive T cells, whose proliferative activity is more robust, are selected for CAR-T cell manufacture ( 180 ). The negative regulators inducing T-cell exhaustion include PD-1, CTLA4, T-cell immunoglobulin and mucin domain 3, and lymphocyte-activation gene 3, which could restrain the activity of T cells while promoting the suppressive function of Tregs ( 181 – 183 ).
The above research advancements may shed light on new strategies to increase CAR-T cell persistence. Engineering strategies to inhibit these negative regulators primarily involve: (1) immune checkpoint blockades, (2) genetic knockdown of negative regulators in CAR-T cells, (3) PD-1 DNR, and (4) autocrine secretion of anti–PD-1 scFv and anti–PD-L1 antibodies from CAR T cells ( 20 , 73 , 182 , 184 ). At present, combination therapy of CAR-T cells and immune checkpoint blockades has been utilized to overcome CAR-T cell exhaustion in clinical trials of NSCLC ( 185 ). CRISPR/Cas9-mediated knockdown of negative regulators in CAR-T cells may become a novel therapeutic approach to increase the persistence of CAR-T cells ( 182 ). CAR-T cells targeting PD-L1z, equipped with CAR-T cells with intrinsic blockade properties of PD-1, demonstrated efficacious antitumor activity in NSCLC models ( 113 ). CAR-T cells secreting anti–PD-L1 antibodies have been demonstrated to combat T cell exhaustion in a renal cell carcinoma mouse model ( 172 ) ( Figure 5 ). In addition, transient cessation of CAR signaling, 4-1BB and CD28 costimulatory signaling, c-Jun, and transcription factors, such as nuclear receptor subfamily 4 group A, NFAT, and thymocyte selection-associated high mobility group box protein have also been shown to regulate T cell exhaustion ( 179 , 184 , 186 ). Further studies are required to apply these findings to enhance CAR-T cell resistance to exhaustion.
3 Future Outlook
CAR-T cell therapy has emerged as a novel and effective immunotherapy against multiple cancers, especially hematological malignancies. The same issues, such as CAR-T therapy-related toxicities, on-target, off-tumor toxicity, and evasion of antitumor responses, have plagued the treatment of hematologic malignancies; the treatment of solid tumors encounters even greater challenges. Moreover, the physical barrier impedes the infiltration of CAR-T cells to tumor sites, and the TME is immunosuppressive. In recent years, the successful improvements in the safety and efficacy of the therapy have facilitated the application of CAR-T therapy in solid tumors, including lung cancer. CAR structures persistently undergo evolution to enhance efficacy and reduce the cytotoxic effects of CAR-T cell therapy. In addition, the engineering solutions mentioned above are in their early stages and are being progressively developed towards the clinical application phase, and further investigations are expected ( Figure 7 ) . Among these engineering strategies, gene editing technology is one of powerful tools to improve the efficacy and safety of CAR-T cell therapy and is driving the application of this novel cancer therapy. The manufacture of “off the shelf” CAR-T cell products by disrupting the TCR alpha/beta chains through TALENs or CRISPR/Cas9 platform, is currently undergoing the evaluation of clinical trials ( 187 ). The inclusion of inducible caspase-9 safety switches to CARs could regulate the production of cytokines to prevent CRS ( 143 ). CRISPR/Cas9-mediated knockdown of negative immune checkpoints enables the CAR-T cells to resist the immunosuppressive TME. It is too early to appreciate the promising prospects of this novel immunotherapy approach in lung cancer treatment until more clinical trials to investigate these engineering strategies are conducted and evaluated.
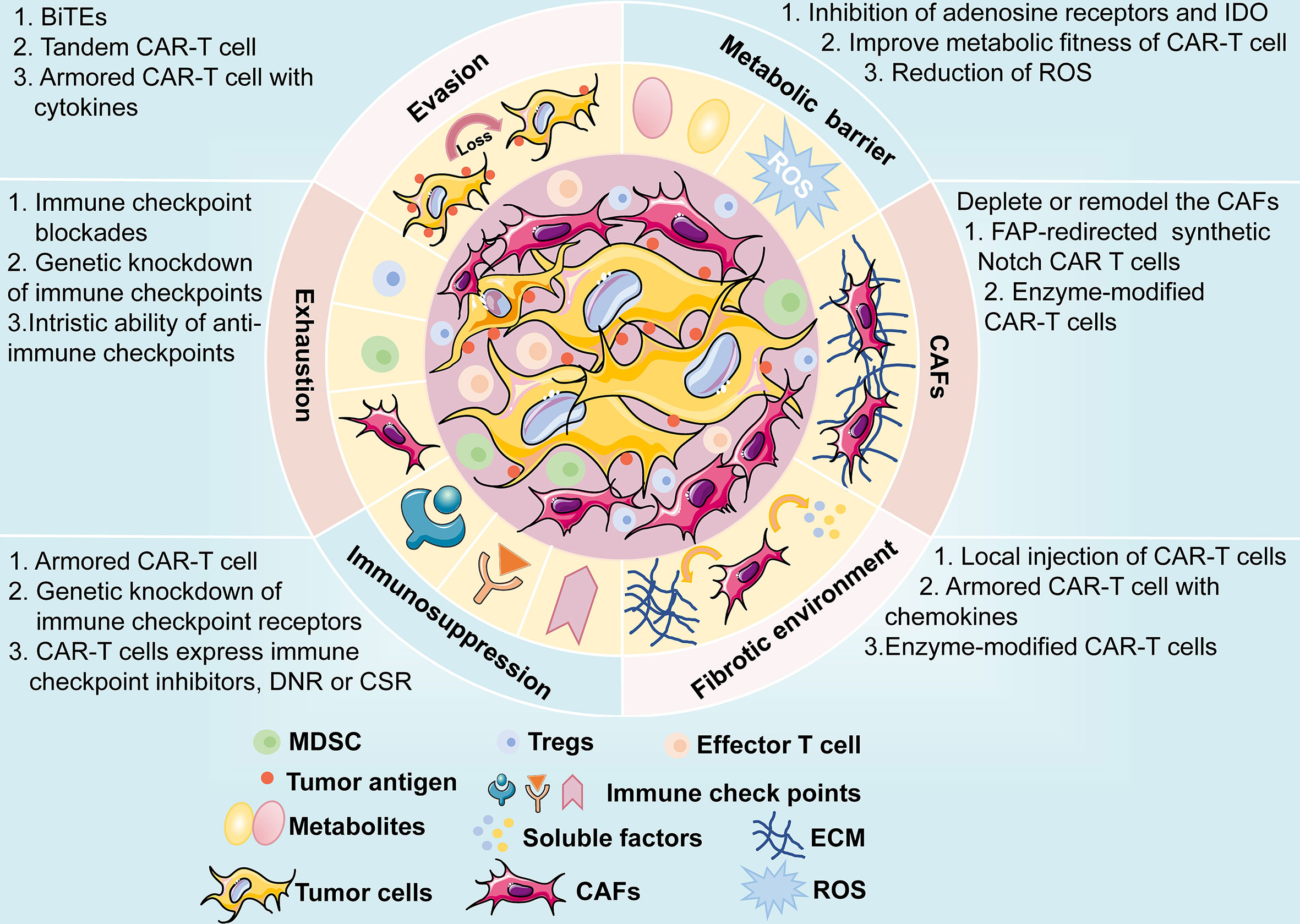
Figure 7 A brief overview of potential challenges faced by CAR-T cell therapy, including antigen evasion, metabolic barrier, CAFs, fibrotic environment, immunosuppression of TME, and exhaustion of CAR-T cells. The possible mechanisms and engineering strategies are also presented.
Author Contributions
B-TY and NW contributed significantly to fund support and the conception of the review. B-FX, J-TZ and Y-GZ contributed to wrote the manuscript. X-RC contributed to make preparations and revise the manuscript. Z-ML helped proposed some constructive suggestions. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Program of China (No. 2018YFC0910700), Beijing Human Resources and Social Security Bureau (Beijing Millions of Talents Project, 2018A05), Beijing Municipal Administration of Hospitals’ Youth Programme (QMS20191107), National Natural Science Foundation of China (No. 81972842), Beijing Natural Science Foundation (No. 7192036), Natural Science Foundation of Jiangxi Province (20202BABL206088).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Wang X, Gong Y, Yao J, Chen Y, Li Y, Zeng Z, et al. Establishment of Criteria for Molecular Differential Diagnosis of MPLC and IPM. Front Oncol (2020) 10:614430. doi: 10.3389/fonc.2020.614430
PubMed Abstract | CrossRef Full Text | Google Scholar
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
3. Niemira M, Collin F, Szalkowska A, Bielska A, Chwialkowska K, Reszec J, et al. Molecular Signature of Subtypes of Non-Small-Cell Lung Cancer by Large-Scale Transcriptional Profiling: Identification of Key Modules and Genes by Weighted Gene Co-Expression Network Analysis (WGCNA). Cancers (2019) 12:37-60. doi: 10.3390/cancers12010037
4. Herbst R, Morgensztern D, Boshoff C. The Biology and Management of Non-Small Cell Lung Cancer. Nature (2018) 553:446–54. doi: 10.1038/nature25183
5. Li J, Zheng Q, Zhao X, Zhao J, An T, Wu M, et al. Nomogram Model for Predicting Cause-Specific Mortality in Patients With Stage I Small-Cell Lung Cancer: A Competing Risk Analysis. BMC Cancer (2020) 20:793. doi: 10.1186/s12885-020-07271-9
6. He S, Lin J, Xu Y, Lin L, Feng J. A Positive Feedback Loop Between ZNF205-AS1 and EGR4 Promotes Non-Small Cell Lung Cancer Growth. J Cell Mol Med (2019) 23:1495–508. doi: 10.1111/jcmm.14056
7. Duma N, Santana-Davila R, Molina JR. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin Proc (2019) 94:1623–40. doi: 10.1016/j.mayocp.2019.01.013
8. Johnson ML, Patel JD. Chemotherapy and Targeted Therapeutics as Maintenance of Response in Advanced Non-Small Cell Lung Cancer. Semin Oncol (2014) 41:93–100. doi: 10.1053/j.seminoncol.2013.12.007
9. Wu SG, Shih JY. Management of Acquired Resistance to EGFR TKI-Targeted Therapy in Advanced Non-Small Cell Lung Cancer. Mol Cancer (2018) 17:38. doi: 10.1186/s12943-018-0777-1
10. Golding B, Luu A, Jones R, Viloria-Petit AM. The Function and Therapeutic Targeting of Anaplastic Lymphoma Kinase (ALK) in Non-Small Cell Lung Cancer (NSCLC). Mol Cancer (2018) 17:52. doi: 10.1186/s12943-018-0810-4
11. Roys A, Chang X, Liu Y, Xu X, Wu Y, Zuo D. Resistance Mechanisms and Potent-Targeted Therapies of ROS1-Positive Lung Cancer. Cancer Chemother Pharmacol (2019) 84:679–88. doi: 10.1007/s00280-019-03902-6
12. Janning M, Loges S. Anti-Angiogenics: Their Value in Lung Cancer Therapy. Oncol Res Treat (2018) 41:172–80. doi: 10.1159/000488119
13. Allaeys T, Berzenji L, Van Schil PE. Surgery After Induction Targeted Therapy and Immunotherapy for Lung Cancer. Cancers (2021) 13:2603–18. doi: 10.3390/cancers13112603
14. Cascone T, William WN, Weissferdt A, Lin HY, Leung CH, Carter BW, et al. Neoadjuvant Nivolumab (N) or Nivolumab Plus Ipilimumab (NI) for Resectable Non-Small Cell Lung Cancer (NSCLC): Clinical and Correlative Results From the NEOSTAR Study. J Clin Oncol (2019) 37:8504–4. doi: 10.1200/JCO.2019.37.15_suppl.8504
CrossRef Full Text | Google Scholar
15. Waqar SN, Morgensztern D. Treatment Advances in Small Cell Lung Cancer (SCLC). Pharmacol Ther (2017) 180:16–23. doi: 10.1016/j.pharmthera.2017.06.002
16. Zhao H, Ren D, Liu H, Chen J. Comparison and Discussion of the Treatment Guidelines for Small Cell Lung Cancer. Thorac Cancer (2018) 9:769–74. doi: 10.1111/1759-7714.12765
17. Yang S, Zhang Z, Wang Q. Emerging Therapies for Small Cell Lung Cancer. J Hematol Oncol (2019) 12:47. doi: 10.1186/s13045-019-0736-3
18. Saltos A, Shafique M, Chiappori A. Update on the Biology, Management, and Treatment of Small Cell Lung Cancer (SCLC). Front Oncol (2020) 10:1074. doi: 10.3389/fonc.2020.01074
19. Barayan R, Ran X, Lok BH. PARP Inhibitors for Small Cell Lung Cancer and Their Potential for Integration Into Current Treatment Approaches. J Thorac Dis (2020) 12:6240–52. doi: 10.21037/jtd.2020.03.89
20. Rafiq S, Hackett CS, Brentjens RJ. Engineering Strategies to Overcome the Current Roadblocks in CAR T Cell Therapy. Nat Rev Clin Oncol (2020) 17:147–67. doi: 10.1038/s41571-019-0297-y
21. Chen N, Li X, Chintala NK, Tano ZE, Adusumilli PS. Driving CARs on the Uneven Road of Antigen Heterogeneity in Solid Tumors. Curr Opin Immunol (2018) 51:103–10. doi: 10.1016/j.coi.2018.03.002
22. Grywalska E, Sosnowska-Pasiarska B, Smok-Kalwat J, Pasiarski M, Niedzwiedzka-Rystwej P, Rolinski J. Paving the Way Toward Successful Multiple Myeloma Treatment: Chimeric Antigen Receptor T-Cell Therapy. Cells (2020) 9:983. doi: 10.3390/cells9040983
23. Stock S, Schmitt M, Sellner L. Optimizing Manufacturing Protocols of Chimeric Antigen Receptor T Cells for Improved Anticancer Immunotherapy. Int J Mol Sci (2019) 20:6223. doi: 10.3390/ijms20246223
24. Han D, Xu Z, Zhuang Y, Ye Z, Qian Q. Current Progress in CAR-T Cell Therapy for Hematological Malignancies. J Cancer (2021) 12:326–34. doi: 10.7150/jca.48976
25. June CH, O'Connor RS, Kawalekar OU, Ghassemi S, Milone MC. CAR T Cell Immunotherapy for Human Cancer. Science (2018) 359:1361–5. doi: 10.1126/science.aar6711
26. Anagnostou T, Riaz IB, Hashmi SK, Murad MH, Kenderian SS. Anti-CD19 Chimeric Antigen Receptor T-Cell Therapy in Acute Lymphocytic Leukaemia: A Systematic Review and Meta-Analysis. Lancet Haematol (2020) 7:e816–26. doi: 10.1016/S2352-3026(20)30277-5
27. Makita S, Imaizumi K, Kurosawa S, Tobinai K. Chimeric Antigen Receptor T-Cell Therapy for B-Cell Non-Hodgkin Lymphoma: Opportunities and Challenges. Drugs Context (2019) 8:212567. doi: 10.7573/dic.212567
28. King AC, Orozco JS. Axicabtagene Ciloleucel: The First FDA-Approved CAR T-Cell Therapy for Relapsed/Refractory Large B-Cell Lymphoma. J Adv Pract Oncol (2019) 10:878–82. doi: 10.6004/jadpro.2019.10.8.9
29. Schuster SJ, Bishop MR, Tam CS, Waller EK, Borchmann P, McGuirk JP, et al. Tisagenlecleucel in Adult Relapsed or Refractory Diffuse Large B-Cell Lymphoma. N Engl J Med (2019) 380:45–56. doi: 10.1056/NEJMoa1804980
30. Maus MV, Alexander S, Bishop MR, Brudno JN, Callahan C, Davila ML, et al. Society for Immunotherapy of Cancer (SITC) Clinical Practice Guideline on Immune Effector Cell-Related Adverse Events. J Immunother Cancer (2020) 8:e001511. doi: 10.1136/jitc-2020-001511
31. Jain P, Nastoupil L, Westin J, Lee HJ, Navsaria L, Steiner RE, et al. Outcomes and Management of Patients With Mantle Cell Lymphoma After Progression on Brexucabtagene Autoleucel Therapy. Br J Haematol (2021) 192:e38–42. doi: 10.1111/bjh.17197
32. Garfall AL, Stadtmauer EA, Hwang WT, Lacey SF, Melenhorst JJ, Krevvata M, et al. Anti-CD19 CAR T Cells With High-Dose Melphalan and Autologous Stem Cell Transplantation for Refractory Multiple Myeloma. JCI Insight (2019) 4:e127684. doi: 10.1172/jci.insight.127684
33. Rodriguez-Lobato LG, Ganzetti M, Fernandez de Larrea C, Hudecek M, Einsele H, Danhof S. CAR T-Cells in Multiple Myeloma: State of the Art and Future Directions. Front Oncol (2020) 10:1243. doi: 10.3389/fonc.2020.01243
34. Ding L, Hu Y, Huang H. Novel Progresses of Chimeric Antigen Receptor (CAR) T Cell Therapy in Multiple Myeloma. Stem Cell Investig (2021) 8:1. doi: 10.21037/sci-2020-029
35. Chen KH, Wada M, Pinz KG, Liu H, Shuai X, Chen X, et al. A Compound Chimeric Antigen Receptor Strategy for Targeting Multiple Myeloma. Leukemia (2018) 32:402–12. doi: 10.1038/leu.2017.302
36. Lin Q, Zhao J, Song Y, Liu D. Recent Updates on CAR T Clinical Trials for Multiple Myeloma. Mol Cancer (2019) 18:154. doi: 10.1186/s12943-019-1092-1
37. Lam N, Trinklein ND, Buelow B, Patterson GH, Ojha N, Kochenderfer JN. Anti-BCMA Chimeric Antigen Receptors With Fully Human Heavy-Chain-Only Antigen Recognition Domains. Nat Commun (2020) 11:283. doi: 10.1038/s41467-019-14119-9
38. Huang R, Li X, He Y, Zhu W, Gao L, Liu Y, et al. Recent Advances in CAR-T Cell Engineering. J Hematol Oncol (2020) 13:86. doi: 10.1186/s13045-020-00910-5
39. Zhang C, Liu J, Zhong JF, Zhang X. Engineering CAR-T Cells. Biomark Res (2017) 5:22. doi: 10.1186/s40364-017-0102-y
40. Yu S, Li A, Liu Q, Li T, Yuan X, Han X, et al. Chimeric Antigen Receptor T Cells: A Novel Therapy for Solid Tumors. J Hematol Oncol (2017) 10:78. doi: 10.1186/s13045-017-0444-9
41. Yeku OO, Purdon TJ, Koneru M, Spriggs D, Brentjens RJ. Armored CAR T Cells Enhance Antitumor Efficacy and Overcome the Tumor Microenvironment. Sci Rep (2017) 7:10541. doi: 10.1038/s41598-017-10940-8
42. Zhang H, Zhao P, Huang H. Engineering Better Chimeric Antigen Receptor T Cells. Exp Hematol Oncol (2020) 9:34. doi: 10.1186/s40164-020-00190-2
43. Singh AP, Zheng X, Lin-Schmidt X, Chen W, Carpenter TJ, Zong A, et al. Development of a Quantitative Relationship Between CAR-Affinity, Antigen Abundance, Tumor Cell Depletion and CAR-T Cell Expansion Using a Multiscale Systems PK-PD Model. MAbs (2020) 12:1688616. doi: 10.1080/19420862.2019.1688616
44. Abreu TR, Fonseca NA, Goncalves N, Moreira JN. Current Challenges and Emerging Opportunities of CAR-T Cell Therapies. J Control Release (2020) 319:246–61. doi: 10.1016/j.jconrel.2019.12.047
45. Fujiwara K, Masutani M, Tachibana M, Okada N. Impact of scFv Structure in Chimeric Antigen Receptor on Receptor Expression Efficiency and Antigen Recognition Properties. Biochem Biophys Res Commun (2020) 527:350–7. doi: 10.1016/j.bbrc.2020.03.071
46. Chang ZL, Lorenzini MH, Chen X, Tran U, Bangayan NJ, Chen YY. Rewiring T-Cell Responses to Soluble Factors With Chimeric Antigen Receptors. Nat Chem Biol (2018) 14:317–24. doi: 10.1038/nchembio.2565
47. Xie YJ, Dougan M, Jailkhani N, Ingram J, Fang T, Kummer L, et al. Nanobody-Based CAR T Cells That Target the Tumor Microenvironment Inhibit the Growth of Solid Tumors in Immunocompetent Mice. Proc Natl Acad Sci USA (2019) 116:7624–31. doi: 10.1073/pnas.1817147116
48. Ingram JR, Schmidt FI, Ploegh HL. Exploiting Nanobodies' Singular Traits. Annu Rev Immunol (2018) 36:695–715. doi: 10.1146/annurev-immunol-042617-053327
49. Mo F, Duan S, Jiang X, Yang X, Hou X, Shi W, et al. Nanobody-Based Chimeric Antigen Receptor T Cells Designed by CRISPR/Cas9 Technology for Solid Tumor Immunotherapy. Signal Transduct Target Ther (2021) 6:80. doi: 10.1038/s41392-021-00462-1
50. Brown CE, Alizadeh D, Starr R, Weng L, Wagner JR, Naranjo A, et al. Regression of Glioblastoma After Chimeric Antigen Receptor T-Cell Therapy. N Engl J Med (2016) 375:2561–9. doi: 10.1056/NEJMoa1610497
51. Baumeister SH, Murad J, Werner L, Daley H, Trebeden-Negre H, Gicobi JK, et al. Phase I Trial of Autologous CAR T Cells Targeting NKG2D Ligands in Patients With AML/MDS and Multiple Myeloma. Cancer Immunol Res (2019) 7:100–12. doi: 10.1158/2326-6066.CIR-18-0307
52. Wang Y, Xu Y, Li S, Liu J, Xing Y, Xing H, et al. Targeting FLT3 in Acute Myeloid Leukemia Using Ligand-Based Chimeric Antigen Receptor-Engineered T Cells. J Hematol Oncol (2018) 11:60. doi: 10.1186/s13045-018-0603-7
53. Lee L, Draper B, Chaplin N, Philip B, Chin M, Galas-Filipowicz D, et al. An APRIL-Based Chimeric Antigen Receptor for Dual Targeting of BCMA and TACI in Multiple Myeloma. Blood (2018) 131:746–58. doi: 10.1182/blood-2017-05-781351
54. Nakazawa Y, Matsuda K, Kurata T, Sueki A, Tanaka M, Sakashita K, et al. Anti-Proliferative Effects of T Cells Expressing a Ligand-Based Chimeric Antigen Receptor Against CD116 on CD34(+) Cells of Juvenile Myelomonocytic Leukemia. J Hematol Oncol (2016) 9:27. doi: 10.1186/s13045-016-0256-3
55. Siegler E, Li S, Kim YJ, Wang P. Designed Ankyrin Repeat Proteins as Her2 Targeting Domains in Chimeric Antigen Receptor-Engineered T Cells. Hum Gene Ther (2017) 28:726–36. doi: 10.1089/hum.2017.021
56. Han X, Cinay GE, Zhao Y, Guo Y, Zhang X, Wang P. Adnectin-Based Design of Chimeric Antigen Receptor for T Cell Engineering. Mol Ther J Am Soc Gene Ther (2017) 25:2466–76. doi: 10.1016/j.ymthe.2017.07.009
57. Fujiwara K, Tsunei A, Kusabuka H, Ogaki E, Tachibana M, Okada N. Hinge and Transmembrane Domains of Chimeric Antigen Receptor Regulate Receptor Expression and Signaling Threshold. Cells (2020) 9:1182. doi: 10.3390/cells9051182
58. Jayaraman J, Mellody MP, Hou AJ, Desai RP, Fung AW, Pham AHT, et al. CAR-T Design: Elements and Their Synergistic Function. EBioMedicine (2020) 58:102931. doi: 10.1016/j.ebiom.2020.102931
59. Hudecek M, Lupo-Stanghellini MT, Kosasih PL, Sommermeyer D, Jensen MC, Rader C, et al. Receptor Affinity and Extracellular Domain Modifications Affect Tumor Recognition by ROR1-Specific Chimeric Antigen Receptor T Cells. Clin Cancer Res (2013) 19:3153–64. doi: 10.1158/1078-0432.CCR-13-0330
60. Schafer D, Henze J, Pfeifer R, Schleicher A, Brauner J, Mockel-Tenbrinck N, et al. A Novel Siglec-4 Derived Spacer Improves the Functionality of CAR T Cells Against Membrane-Proximal Epitopes. Front Immunol (2020) 11:1704. doi: 10.3389/fimmu.2020.01704
61. Hudecek M, Sommermeyer D, Kosasih PL, Silva-Benedict A, Liu L, Rader C, et al. The Nonsignaling Extracellular Spacer Domain of Chimeric Antigen Receptors Is Decisive for In Vivo Antitumor Activity. Cancer Immunol Res (2015) 3:125–35. doi: 10.1158/2326-6066.CIR-14-0127
62. Guedan S, Posey AD Jr, Shaw C, Wing A, Da T, Patel PR, et al. Enhancing CAR T Cell Persistence Through ICOS and 4-1BB Costimulation. JCI Insight (2018) 3:e96976. doi: 10.1172/jci.insight.96976
63. Bridgeman JS, Hawkins RE, Bagley S, Blaylock M, Holland M, Gilham DE. The Optimal Antigen Response of Chimeric Antigen Receptors Harboring the CD3zeta Transmembrane Domain Is Dependent Upon Incorporation of the Receptor Into the Endogenous TCR/CD3 Complex. J Immunol (Baltimore Md 1950) (2010) 184:6938–49. doi: 10.4049/jimmunol.0901766
64. Ying Z, Huang XF, Xiang X, Liu Y, Kang X, Song Y, et al. A Safe and Potent Anti-CD19 CAR T Cell Therapy. Nat Med (2019) 25:947–53. doi: 10.1038/s41591-019-0421-7
65. Wan Z, Shao X, Ji X, Dong L, Wei J, Xiong Z, et al. Transmembrane Domain-Mediated Lck Association Underlies Bystander and Costimulatory ICOS Signaling. Cell Mol Immunol (2020) 17:143–52. doi: 10.1038/s41423-018-0183-z
66. Weinkove R, George P, Dasyam N, McLellan AD. Selecting Costimulatory Domains for Chimeric Antigen Receptors: Functional and Clinical Considerations. Clin Trans Immunol (2019) 8:e1049. doi: 10.1002/cti2.1049
67. Qu J, Mei Q, Chen L, Zhou J. Chimeric Antigen Receptor (CAR)-T-Cell Therapy in Non-Small-Cell Lung Cancer (NSCLC): Current Status and Future Perspectives. Cancer Immunol Immunother (2021) 70:619–31. doi: 10.1007/s00262-020-02735-0
68. Chandran SS, Klebanoff CA. T Cell Receptor-Based Cancer Immunotherapy: Emerging Efficacy and Pathways of Resistance. Immunol Rev (2019) 290:127–47. doi: 10.1111/imr.12772
69. van der Stegen SJ, Hamieh M, Sadelain M. The Pharmacology of Second-Generation Chimeric Antigen Receptors. Nat Rev Drug Discov (2015) 14:499–509. doi: 10.1038/nrd4597
70. Long AH, Haso WM, Shern JF, Wanhainen KM, Murgai M, Ingaramo M, et al. 4-1BB Costimulation Ameliorates T Cell Exhaustion Induced by Tonic Signaling of Chimeric Antigen Receptors. Nat Med (2015) 21:581–90. doi: 10.1038/nm.3838
71. Philipson BI, O'Connor RS, May MJ, June CH, Albelda SM, Milone MC. 4-1BB Costimulation Promotes CAR T Cell Survival Through Noncanonical NF-KappaB Signaling. Sci Signal (2020) 13:e8248. doi: 10.1126/scisignal.aay8248
72. Sun C, Shou P, Du H, Hirabayashi K, Chen Y, Herring LE, et al. THEMIS-SHP1 Recruitment by 4-1BB Tunes LCK-Mediated Priming of Chimeric Antigen Receptor-Redirected T Cells. Cancer Cell (2020) 37:216–225 e6. doi: 10.1016/j.ccell.2019.12.014
73. Larson RC, Maus MV. Recent Advances and Discoveries in the Mechanisms and Functions of CAR T Cells. Nat Rev Cancer (2021) 21:145–61. doi: 10.1038/s41568-020-00323-z
74. Song DG, Powell DJ. Pro-Survival Signaling via CD27 Costimulation Drives Effective CAR T-Cell Therapy. Oncoimmunology (2012) 1:547–9. doi: 10.4161/onci.19458
75. Duong CP, Westwood JA, Yong CS, Murphy A, Devaud C, John LB, et al. Engineering T Cell Function Using Chimeric Antigen Receptors Identified Using a DNA Library Approach. PloS One (2013) 8:e63037. doi: 10.1371/journal.pone.0063037
76. Hombach AA, Chmielewski M, Rappl G, Abken H. Adoptive Immunotherapy With Redirected T Cells Produces CCR7- Cells That Are Trapped in the Periphery and Benefit From Combined CD28-OX40 Costimulation. Hum Gene Ther (2013) 24:259–69. doi: 10.1089/hum.2012.247
77. Hombach AA, Heiders J, Foppe M, Chmielewski M, Abken H. OX40 Costimulation by a Chimeric Antigen Receptor Abrogates CD28 and IL-2 Induced IL-10 Secretion by Redirected CD4(+) T Cells. Oncoimmunology (2012) 1:458–66. doi: 10.4161/onci.19855
78. Mata M, Gerken C, Nguyen P, Krenciute G, Spencer DM, Gottschalk S. Inducible Activation of MyD88 and CD40 in CAR T Cells Results in Controllable and Potent Antitumor Activity in Preclinical Solid Tumor Models. Cancer Discov (2017) 7:1306–19. doi: 10.1158/2159-8290.CD-17-0263
79. Wang E, Wang LC, Tsai CY, Bhoj V, Gershenson Z, Moon E, et al. Generation of Potent T-Cell Immunotherapy for Cancer Using DAP12-Based, Multichain, Chimeric Immunoreceptors. Cancer Immunol Res (2015) 3:815–26. doi: 10.1158/2326-6066.CIR-15-0054
80. Singh AK, McGuirk JP. CAR T Cells: Continuation in a Revolution of Immunotherapy. Lancet Oncol (2020) 21:e168–78. doi: 10.1016/S1470-2045(19)30823-X
81. van der Stegen S, Hamieh M, Sadelain M. The Pharmacology of Second-Generation Chimeric Antigen Receptors. Nat Rev Drug Discov (2015) 14:499–509. doi: 10.1038/nrd4597
82. D'Aloia MM, Zizzari IG, Sacchetti B, Pierelli L, Alimandi M. CAR-T Cells: The Long and Winding Road to Solid Tumors. Cell Death Dis (2018) 9:282. doi: 10.1038/s41419-018-0278-6
83. Guedan S, Ruella M, June CH. Emerging Cellular Therapies for Cancer. Annu Rev Immunol (2019) 37:145–71. doi: 10.1146/annurev-immunol-042718-041407
84. Chmielewski M, Abken H. TRUCKs: The Fourth Generation of CARs. Expert Opin Biol Ther (2015) 15:1145–54. doi: 10.1517/14712598.2015.1046430
85. Zhao L, Cao YJ. Engineered T Cell Therapy for Cancer in the Clinic. Front Immunol (2019) 10:2250. doi: 10.3389/fimmu.2019.02250
86. Zhong S, Cui Y, Liu Q, Chen S. CAR-T Cell Therapy for Lung Cancer: A Promising But Challenging Future. J Thorac Dis (2020) 12:4516–21. doi: 10.21037/jtd.2020.03.118
87. Klampatsa A, Dimou V, Albelda SM. Mesothelin-Targeted CAR-T Cell Therapy for Solid Tumors. Expert Opin Biol Ther (2021) 21:473–86. doi: 10.1080/14712598.2021.1843628
88. Hagemann UB, Ellingsen C, Schuhmacher J, Kristian A, Mobergslien A, Cruciani V, et al. Mesothelin-Targeted Thorium-227 Conjugate (MSLN-TTC): Preclinical Evaluation of a New Targeted Alpha Therapy for Mesothelin-Positive Cancers. Clin Cancer Res (2019) 25:4723–34. doi: 10.1158/1078-0432.CCR-18-3476
89. Ye L, Lou Y, Lu L, Fan X. Mesothelin-Targeted Second Generation CAR-T Cells Inhibit Growth of Mesothelin-Expressing Tumors In Vivo . Exp Ther Med (2019) 17:739–47. doi: 10.3892/etm.2018.7015
90. Morello A, Sadelain M, Adusumilli PS. Mesothelin-Targeted CARs: Driving T Cells to Solid Tumors. Cancer Discov (2016) 6:133–46. doi: 10.1158/2159-8290.CD-15-0583
91. da Cunha Santos G, Shepherd FA, Tsao MS. EGFR Mutations and Lung Cancer. Annu Rev Pathol (2011) 6:49–69. doi: 10.1146/annurev-pathol-011110-130206
92. Zhang Z, Jiang J, Wu X, Zhang M, Luo D, Zhang R, et al. Chimeric Antigen Receptor T Cell Targeting EGFRvIII for Metastatic Lung Cancer Therapy. Front Med (2019) 13:57–68. doi: 10.1007/s11684-019-0683-y
93. Li H, Huang Y, Jiang DQ, Cui LZ, He Z, Wang C, et al. Antitumor Activity of EGFR-Specific CAR T Cells Against Non-Small-Cell Lung Cancer Cells In Vitro and in Mice. Cell Death Dis (2018) 9:177. doi: 10.1038/s41419-017-0238-6
94. Liu D. CAR-T "the Living Drugs", Immune Checkpoint Inhibitors, and Precision Medicine: A New Era of Cancer Therapy. J Hematol Oncol (2019) 12:113. doi: 10.1186/s13045-019-0819-1
95. Yamaguchi T, Yanagisawa K, Sugiyama R, Hosono Y, Shimada Y, Arima C, et al. NKX2-1/TITF1/TTF-1-Induced ROR1 Is Required to Sustain EGFR Survival Signaling in Lung Adenocarcinoma. Cancer Cell (2012) 21:348–61. doi: 10.1016/j.ccr.2012.02.008
96. Schiavone G, Epistolio S, Martin V, Molinari F, Barizzi J, Mazzucchelli L, et al. Functional and Clinical Significance of ROR1 in Lung Adenocarcinoma. BMC Cancer (2020) 20:1085. doi: 10.1186/s12885-020-07587-6
97. Wallstabe L, Gottlich C, Nelke LC, Kuhnemundt J, Schwarz T, Nerreter T, et al. ROR1-CAR T Cells Are Effective Against Lung and Breast Cancer in Advanced Microphysiologic 3D Tumor Models. JCI Insight (2019) 4:e126345. doi: 10.1172/jci.insight.126345
98. Balakrishnan A, Goodpaster T, Randolph-Habecker J, Hoffstrom BG, Jalikis FG, Koch LK, et al. Analysis of ROR1 Protein Expression in Human Cancer and Normal Tissues. Clin Cancer Res (2017) 23:3061–71. doi: 10.1158/1078-0432.CCR-16-2083
99. Srivastava S, Furlan SN, Jaeger-Ruckstuhl CA, Sarvothama M, Berger C, Smythe KS, et al. Immunogenic Chemotherapy Enhances Recruitment of CAR-T Cells to Lung Tumors and Improves Antitumor Efficacy When Combined With Checkpoint Blockade. Cancer Cell (2021) 39:193–208.e10. doi: 10.1016/j.ccell.2020.11.005
100. Berger C, Sommermeyer D, Hudecek M, Berger M, Balakrishnan A, Paszkiewicz PJ, et al. Safety of Targeting ROR1 in Primates With Chimeric Antigen Receptor-Modified T Cells. Cancer Immunol Res (2015) 3:206–16. doi: 10.1158/2326-6066.Cir-14-0163
101. Bouillez A, Adeegbe D, Jin C, Hu X, Tagde A, Alam M, et al. MUC1-C Promotes the Suppressive Immune Microenvironment in Non-Small Cell Lung Cancer. Oncoimmunology (2017) 6:e1338998. doi: 10.1080/2162402X.2017.1338998
102. Jiang ZB, Huang JM, Xie YJ, Zhang YZ, Chang C, Lai HL, et al. Evodiamine Suppresses Non-Small Cell Lung Cancer by Elevating CD8(+) T Cells and Downregulating the MUC1-C/PD-L1 Axis. J Exp Clin Cancer Res (2020) 39:249. doi: 10.1186/s13046-020-01741-5
103. Saeki N, Gu J, Yoshida T, Wu X. Prostate Stem Cell Antigen: A Jekyll and Hyde Molecule? Clin Cancer Res (2010) 16:3533–8. doi: 10.1158/1078-0432.Ccr-09-3169
104. Wei X, Lai Y, Li J, Qin L, Xu Y, Zhao R, et al. PSCA and MUC1 in Non-Small-Cell Lung Cancer as Targets of Chimeric Antigen Receptor T Cells. Oncoimmunology (2017) 6:e1284722. doi: 10.1080/2162402x.2017.1284722
105. Soliman AM, Alqahtani AS, Ghorab M. Novel Sulphonamide Benzoquinazolinones as Dual EGFR/HER2 Inhibitors, Apoptosis Inducers and Radiosensitizers. J Enzyme Inhib Med Chem (2019) 34:1030–40. doi: 10.1080/14756366.2019.1609469
106. McKenna MK, Englisch A, Brenner B, Smith T, Hoyos V, Suzuki M, et al. Mesenchymal Stromal Cell Delivery of Oncolytic Immunotherapy Improves CAR-T Cell Antitumor Activity. Mol Ther J Am Soc Gene Ther (2021) 29:1808–20. doi: 10.1016/j.ymthe.2021.02.004
107. Gao Q, Wang S, Chen X, Cheng S, Zhang Z, Li F, et al. Cancer-Cell-Secreted CXCL11 Promoted CD8(+) T Cells Infiltration Through Docetaxel-Induced-Release of HMGB1 in NSCLC. J Immunother Cancer (2019) 7:42. doi: 10.1186/s40425-019-0511-6
108. Zeltsman M, Dozier J, McGee E, Ngai D, Adusumilli PS. CAR T-Cell Therapy for Lung Cancer and Malignant Pleural Mesothelioma. Trans Res J Lab Clin Med (2017) 187:1–10. doi: 10.1016/j.trsl.2017.04.004
109. Li X, Liu M, Zhang H, Liu H, Chen J. Clinical Study of Apatinib Combined With EGFR-TKI in the Treatment of Chronic Progression After EGFR-TKI Treatment in Non-Small Cell Lung Cancer (Chictr1800019185). Thorac Cancer (2020) 11:819–26. doi: 10.1111/1759-7714.13303
110. Chmielewski M, Abken H. CAR T Cells Releasing IL-18 Convert to T-Bet(high) FoxO1(low) Effectors That Exhibit Augmented Activity Against Advanced Solid Tumors. Cell Rep (2017) 21:3205–19. doi: 10.1016/j.celrep.2017.11.063
111. Chen Y, Pei Y, Luo J, Huang Z, Yu J, Meng X. Looking for the Optimal PD-1/PD-L1 Inhibitor in Cancer Treatment: A Comparison in Basic Structure, Function, and Clinical Practice. Front Immunol (2020) 11:1088. doi: 10.3389/fimmu.2020.01088
112. Liu M, Wang X, Li W, Yu X, Flores-Villanueva P, Xu-Monette ZY, et al. Targeting PD-L1 in Non-Small Cell Lung Cancer Using CAR T Cells. Oncogenesis (2020) 9:72. doi: 10.1038/s41389-020-00257-z
113. Qin L, Zhao R, Chen D, Wei X, Wu Q, Long Y, et al. Chimeric Antigen Receptor T Cells Targeting PD-L1 Suppress Tumor Growth. Biomark Res (2020) 8:19. doi: 10.1186/s40364-020-00198-0
114. Kakarla S, Chow KK, Mata M, Shaffer DR, Song XT, Wu MF, et al. Antitumor Effects of Chimeric Receptor Engineered Human T Cells Directed to Tumor Stroma. Mol Ther J Am Soc Gene Ther (2013) 21:1611–20. doi: 10.1038/mt.2013.110
115. Busek P, Mateu R, Zubal M, Kotackova L, Sedo A. Targeting Fibroblast Activation Protein in Cancer - Prospects and Caveats. Front Biosci (Landmark Ed) (2018) 23:1933–68. doi: 10.2741/4682
116. Wang LC, Lo A, Scholler J, Sun J, Majumdar RS, Kapoor V, et al. Targeting Fibroblast Activation Protein in Tumor Stroma With Chimeric Antigen Receptor T Cells can Inhibit Tumor Growth and Augment Host Immunity Without Severe Toxicity. Cancer Immunol Res (2014) 2:154–66. doi: 10.1158/2326-6066.CIR-13-0027
117. Tran E, Chinnasamy D, Yu Z, Morgan RA, Lee CC, Restifo NP, et al. Immune Targeting of Fibroblast Activation Protein Triggers Recognition of Multipotent Bone Marrow Stromal Cells and Cachexia. J Exp Med (2013) 210:1125–35. doi: 10.1084/jem.20130110
118. Hu Z, Zheng X, Jiao D, Zhou Y, Sun R, Wang B, et al. LunX-CAR T Cells as a Targeted Therapy for Non-Small Cell Lung Cancer. Mol Ther Oncolytics (2020) 17:361–70. doi: 10.1016/j.omto.2020.04.008
119. Porcellini S, Asperti C, Corna S, Cicoria E, Valtolina V, Stornaiuolo A, et al. CAR T Cells Redirected to CD44v6 Control Tumor Growth in Lung and Ovary Adenocarcinoma Bearing Mice. Front Immunol (2020) 11:99. doi: 10.3389/fimmu.2020.00099
120. Mao Y, Fan W, Hu H, Zhang L, Michel J, Wu Y, et al. MAGE-A1 in Lung Adenocarcinoma as a Promising Target of Chimeric Antigen Receptor T Cells. J Hematol Oncol (2019) 12:106. doi: 10.1186/s13045-019-0793-7
121. Li N, Liu S, Sun M, Chen W, Xu X, Zeng Z, et al. Chimeric Antigen Receptor-Modified T Cells Redirected to EphA2 for the Immunotherapy of Non-Small Cell Lung Cancer. Trans Oncol (2018) 11:11–7. doi: 10.1016/j.tranon.2017.10.009
122. Shimizu Y, Suzuki T, Yoshikawa T, Endo I, Nakatsura T. Next-Generation Cancer Immunotherapy Targeting Glypican-3. Front Oncol (2019) 9:248. doi: 10.3389/fonc.2019.00248
123. Crossland DL, Denning WL, Ang S, Olivares S, Mi T, Switzer K, et al. Antitumor Activity of CD56-Chimeric Antigen Receptor T Cells in Neuroblastoma and SCLC Models. Oncogene (2018) 37:3686–97. doi: 10.1038/s41388-018-0187-2
124. Owen DH, Giffin MJ, Bailis JM, Smit MD, Carbone DP, He K. DLL3: An Emerging Target in Small Cell Lung Cancer. J Hematol Oncol (2019) 12:61. doi: 10.1186/s13045-019-0745-2
125. Yang M, Tang X, Zhang Z, Gu L, Wei H, Zhao S, et al. Tandem CAR-T Cells Targeting CD70 and B7-H3 Exhibit Potent Preclinical Activity Against Multiple Solid Tumors. Theranostics (2020) 10:7622–34. doi: 10.7150/thno.43991
126. Pardoll DM. The Blockade of Immune Checkpoints in Cancer Immunotherapy. Nat Rev Cancer (2012) 12:252–64. doi: 10.1038/nrc3239
127. Kontos F, Michelakos T, Kurokawa T, Sadagopan A, Schwab JH, Ferrone CR, et al. B7-H3: An Attractive Target for Antibody-Based Immunotherapy. Clin Cancer Res (2021) 27:1227–35. doi: 10.1158/1078-0432.CCR-20-2584
128. Lin S, Cheng L, Ye W, Li S, Zheng D, Qin L, et al. Chimeric CTLA4-CD28-CD3z T Cells Potentiate Antitumor Activity Against CD80/CD86-Positive B Cell Malignancies. Front Immunol (2021) 12:642528. doi: 10.3389/fimmu.2021.642528
129. Chu W, Zhou Y, Tang Q, Wang M, Ji Y, Yan J, et al. Bi-Specific Ligand-Controlled Chimeric Antigen Receptor T-Cell Therapy for Non-Small Cell Lung Cancer. Biosci Trends (2018) 12:298–308. doi: 10.5582/bst.2018.01048
130. Springuel L, Lonez C, Alexandre B, Van Cutsem E, Machiels JH, Van Den Eynde M, et al. Chimeric Antigen Receptor-T Cells for Targeting Solid Tumors: Current Challenges and Existing Strategies. BioDrugs (2019) 33:515–37. doi: 10.1007/s40259-019-00368-z
131. Kiesgen S, Chicaybam L, Chintala NK, Adusumilli PS. Chimeric Antigen Receptor (CAR) T-Cell Therapy for Thoracic Malignancies. J Thorac Oncol (2018) 13(1):16–26. doi: 10.1016/j.jtho.2018.01.001
132. Morgan RA, Yang JC, Kitano M, Dudley ME, Laurencot CM, Rosenberg SA. Case Report of a Serious Adverse Event Following the Administration of T Cells Transduced With a Chimeric Antigen Receptor Recognizing ERBB2. Mol Ther J Am Soc Gene Ther (2010) 18:843–51. doi: 10.1038/mt.2010.24
133. Morris EC, Neelapu SS, Giavridis T, Sadelain M. Cytokine Release Syndrome and Associated Neurotoxicity in Cancer Immunotherapy. Nat Rev Immunol (2021) 1–12. doi: 10.1038/s41577-021-00547-6
134. Kasakovski D, Xu L, Li Y. T Cell Senescence and CAR-T Cell Exhaustion in Hematological Malignancies. J Hematol Oncol (2018) 11:91. doi: 10.1186/s13045-018-0629-x
135. Hou AJ, Chen LC, Chen YY. Navigating CAR-T Cells Through the Solid-Tumour Microenvironment. Nat Rev Drug Discov (2021) 20(7):531–50. doi: 10.1038/s41573-021-00189-2
136. Frey N, Porter D. Cytokine Release Syndrome With Chimeric Antigen Receptor T Cell Therapy. Biol Blood Marrow Transplant (2019) 25:e123–7. doi: 10.1016/j.bbmt.2018.12.756
137. Shimabukuro-Vornhagen A, Gödel P, Subklewe M, Stemmler HJ, Schlößer HA, Schlaak M, et al. Cytokine Release Syndrome. J Immunother Cancer (2018) 6:56. doi: 10.1186/s40425-018-0343-9
138. Lee DW, Santomasso BD, Locke FL, Ghobadi A, Turtle CJ, Brudno JN, et al. ASTCT Consensus Grading for Cytokine Release Syndrome and Neurologic Toxicity Associated With Immune Effector Cells. Biol Blood Marrow Transplant (2019) 25:625–38. doi: 10.1016/j.bbmt.2018.12.758
139. Rivera AM, May S, Lei M, Qualls S, Bushey K, Rubin DB, et al. CAR T-Cell-Associated Neurotoxicity: Current Management and Emerging Treatment Strategies. Crit Care Nurs Q (2020) 43:191–204. doi: 10.1097/CNQ.0000000000000302
140. Carter SJ, Tattersall RS, Ramanan AV. Macrophage Activation Syndrome in Adults: Recent Advances in Pathophysiology, Diagnosis and Treatment. Rheumatology (Oxford) (2019) 58:5–17. doi: 10.1093/rheumatology/key006
141. Neelapu SS, Tummala S, Kebriaei P, Wierda W, Gutierrez C, Locke FL, et al. Chimeric Antigen Receptor T-Cell Therapy — Assessment and Management of Toxicities. Nat Rev Clin Oncol (2018) 15:47–62. doi: 10.1038/nrclinonc.2017.148
142. Salter AI, Ivey RG, Kennedy JJ, Voillet V, Rajan A, Alderman EJ, et al. Phosphoproteomic Analysis of Chimeric Antigen Receptor Signaling Reveals Kinetic and Quantitative Differences That Affect Cell Function. Sci Signal (2018) 11:eaat6753. doi: 10.1126/scisignal.aat6753
143. Diaconu I, Ballard B, Zhang M, Chen Y, West J, Dotti G, et al. Inducible Caspase-9 Selectively Modulates the Toxicities of CD19-Specific Chimeric Antigen Receptor-Modified T Cells. Mol Ther J Am Soc Gene Ther (2017) 25:580–92. doi: 10.1016/j.ymthe.2017.01.011
144. Liu J, Yang S, Cao B, Zhou G, Zhang F, Wang Y, et al. Targeting B7-H3 via Chimeric Antigen Receptor T Cells and Bispecific Killer Cell Engagers Augments Antitumor Response of Cytotoxic Lymphocytes. J Hematol Oncol (2021) 14:21. doi: 10.1186/s13045-020-01024-8
145. Lamers CH, Sleijfer S, van Steenbergen S, van Elzakker P, van Krimpen B, Groot C, et al. Treatment of Metastatic Renal Cell Carcinoma With CAIX CAR-Engineered T Cells: Clinical Evaluation and Management of on-Target Toxicity. Mol Ther J Am Soc Gene Ther (2013) 21:904–12. doi: 10.1038/mt.2013.17
146. Yu S, Yi M, Qin S, Wu K. Next Generation Chimeric Antigen Receptor T Cells: Safety Strategies to Overcome Toxicity. Mol Cancer (2019) 18:125. doi: 10.1186/s12943-019-1057-4
147. Wu Y, Liu Y, Huang Z, Wang X, Jin Z, Li J, et al. Control of the Activity of CAR-T Cells Within Tumours via Focused Ultrasound. Nat Biomed Eng (2021). doi: 10.1038/s41551-021-00779-w
148. Song DG, Ye Q, Poussin M, Chacon JA, Figini M, Powell DJ Jr. Effective Adoptive Immunotherapy of Triple-Negative Breast Cancer by Folate Receptor-Alpha Redirected CAR T Cells Is Influenced by Surface Antigen Expression Level. J Hematol Oncol (2016) 9:56. doi: 10.1186/s13045-016-0285-y
149. Tong C, Zhang Y, Liu Y, Ji X, Zhang W, Guo Y, et al. Optimized Tandem CD19/CD20 CAR-Engineered T Cells in Refractory/Relapsed B-Cell Lymphoma. Blood (2020) 136:1632–44. doi: 10.1182/blood.2020005278
150. Majzner RG, Mackall CL. Tumor Antigen Escape From CAR T-Cell Therapy. Cancer Discov (2018) 8:1219–26. doi: 10.1158/2159-8290.CD-18-0442
151. Caswell DR, Swanton C. The Role of Tumour Heterogeneity and Clonal Cooperativity in Metastasis, Immune Evasion and Clinical Outcome. BMC Med (2017) 15:133. doi: 10.1186/s12916-017-0900-y
152. O'Rourke DM, Nasrallah MP, Desai A, Melenhorst JJ, Mansfield K, Morrissette JJD, et al. A Single Dose of Peripherally Infused EGFRvIII-Directed CAR T Cells Mediates Antigen Loss and Induces Adaptive Resistance in Patients With Recurrent Glioblastoma. Sci Transl Med (2017) 9:eaaa0984. doi: 10.1126/scitranslmed.aaa0984
153. Rafiq S, Brentjens RJ. Tumors Evading CARs-The Chase Is On. Nat Med (2018) 24:1492–3. doi: 10.1038/s41591-018-0212-6
154. Choi BD, Yu X, Castano AP, Bouffard AA, Schmidts A, Larson RC, et al. CAR-T Cells Secreting BiTEs Circumvent Antigen Escape Without Detectable Toxicity. Nat Biotechnol (2019) 37:1049–58. doi: 10.1038/s41587-019-0192-1
155. Goebeler M-E, Bargou RC. T Cell-Engaging Therapies — BiTEs and Beyond. Nat Rev Clin Oncol (2020) 17:418–34. doi: 10.1038/s41571-020-0347-5
156. Hegde M, Mukherjee M, Grada Z, Pignata A, Landi D, Navai SA, et al. Tandem CAR T Cells Targeting HER2 and IL13Rα2 Mitigate Tumor Antigen Escape. J Clin Invest (2016) 126:3036–52. doi: 10.1172/jci83416
157. Grada Z, Hegde M, Byrd T, Shaffer DR, Ghazi A, Brawley VS, et al. TanCAR: A Novel Bispecific Chimeric Antigen Receptor for Cancer Immunotherapy. Mol Ther Nucleic Acids (2013) 2:e105. doi: 10.1038/mtna.2013.32
158. Avanzi MP, Yeku O, Li X, Wijewarnasuriya DP, van Leeuwen DG, Cheung K, et al. Engineered Tumor-Targeted T Cells Mediate Enhanced Anti-Tumor Efficacy Both Directly and Through Activation of the Endogenous Immune System. Cell Rep (2018) 23:2130–41. doi: 10.1016/j.celrep.2018.04.051
159. Nurmik M, Ullmann P, Rodriguez F, Haan S, Letellier E. In Search of Definitions: Cancer-Associated Fibroblasts and Their Markers. Int J Cancer (2020) 146:895–905. doi: 10.1002/ijc.32193
160. Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, et al. A Framework for Advancing Our Understanding of Cancer-Associated Fibroblasts. Nat Rev Cancer (2020) 20:174–86. doi: 10.1038/s41568-019-0238-1
161. Biffi G, Tuveson DA. Diversity and Biology of Cancer-Associated Fibroblasts. Physiol Rev (2021) 101:147–76. doi: 10.1152/physrev.00048.2019
162. Kobayashi H, Enomoto A, Woods SL, Burt AD, Takahashi M, Worthley DL. Cancer-Associated Fibroblasts in Gastrointestinal Cancer. Nat Rev Gastroenterol Hepatol (2019) 16:282–95. doi: 10.1038/s41575-019-0115-0
163. Henze J, Tacke F, Hardt O, Alves F, Al Rawashdeh W. Enhancing the Efficacy of CAR T Cells in the Tumor Microenvironment of Pancreatic Cancer. Cancers (2020) 12:1389–409. doi: 10.3390/cancers12061389
164. Valkenburg KC, de Groot AE, Pienta KJ. Targeting the Tumour Stroma to Improve Cancer Therapy. Nat Rev Clin Oncol (2018) 15:366–81. doi: 10.1038/s41571-018-0007-1
165. Salmon H, Franciszkiewicz K, Damotte D, Dieu-Nosjean MC, Validire P, Trautmann A, et al. Matrix Architecture Defines the Preferential Localization and Migration of T Cells Into the Stroma of Human Lung Tumors. J Clin Invest (2012) 122:899–910. doi: 10.1172/JCI45817
166. Jiang H, Hegde S, DeNardo DG. Tumor-Associated Fibrosis as a Regulator of Tumor Immunity and Response to Immunotherapy. Cancer Immunol Immunother (2017) 66:1037–48. doi: 10.1007/s00262-017-2003-1
167. Craddock JA, Lu A, Bear A, Pule M, Brenner MK, Rooney CM, et al. Enhanced Tumor Trafficking of GD2 Chimeric Antigen Receptor T Cells by Expression of the Chemokine Receptor CCR2b. J Immunother (Hagerstown Md 1997) (2010) 33:780–8. doi: 10.1097/CJI.0b013e3181ee6675
168. Adachi K, Kano Y, Nagai T, Okuyama N, Sakoda Y, Tamada K. IL-7 and CCL19 Expression in CAR-T Cells Improves Immune Cell Infiltration and CAR-T Cell Survival in the Tumor. Nat Biotechnol (2018) 36:346–51. doi: 10.1038/nbt.4086
169. Caruana I, Savoldo B, Hoyos V, Weber G, Liu H, Kim ES, et al. Heparanase Promotes Tumor Infiltration and Antitumor Activity of CAR-Redirected T Lymphocytes. Nat Med (2015) 21:524–9. doi: 10.1038/nm.3833
170. Dragon AC, Zimmermann K, Nerreter T, Sandfort D, Lahrberg J, Kloss S, et al. CAR-T Cells and TRUCKs That Recognize an EBNA-3C-Derived Epitope Presented on HLA-B*35 Control Epstein-Barr Virus-Associated Lymphoproliferation. J Immunother Cancer (2020) 8:e000736. doi: 10.1136/jitc-2020-000736
171. McGowan E, Lin Q, Ma G, Yin H, Chen S, Lin Y. PD-1 Disrupted CAR-T Cells in the Treatment of Solid Tumors: Promises and Challenges. BioMed Pharmacother (2020) 121:109625. doi: 10.1016/j.biopha.2019.109625
172. Suarez ER, Chang de K, Sun J, Sui J, Freeman GJ, Signoretti S, et al. Chimeric Antigen Receptor T Cells Secreting Anti-PD-L1 Antibodies More Effectively Regress Renal Cell Carcinoma in a Humanized Mouse Model. Oncotarget (2016) 7:34341–55. doi: 10.18632/oncotarget.9114
173. Tay JC, Zha S, Wang S. Chimeric Switch Receptor: Switching for Improved Adoptive T-Cell Therapy Against Cancers. Immunotherapy (2017) 9:1339–49. doi: 10.2217/imt-2017-0103
174. Beckermann KE, Dudzinski SO, Rathmell JC. Dysfunctional T Cell Metabolism in the Tumor Microenvironment. Cytokine Growth Factor Rev (2017) 35:7–14. doi: 10.1016/j.cytogfr.2017.04.003
175. Newick K, O'Brien S, Moon E, Albelda SM. CAR T Cell Therapy for Solid Tumors. Annu Rev Med (2017) 68:139–52. doi: 10.1146/annurev-med-062315-120245
176. Beavis PA, Henderson MA, Giuffrida L, Mills JK, Sek K, Cross RS, et al. Targeting the Adenosine 2A Receptor Enhances Chimeric Antigen Receptor T Cell Efficacy. J Clin Invest (2017) 127:929–41. doi: 10.1172/JCI89455
177. Pellegrino M, Del Bufalo F, De Angelis B, Quintarelli C, Caruana I, de Billy E. Manipulating the Metabolism to Improve the Efficacy of CAR T-Cell Immunotherapy. Cells (2020) 10:14-29 doi: 10.3390/cells10010014
178. Menk AV, Scharping NE, Rivadeneira DB, Calderon MJ, Watson MJ, Dunstane D, et al. 4-1BB Costimulation Induces T Cell Mitochondrial Function and Biogenesis Enabling Cancer Immunotherapeutic Responses. J Exp Med (2018) 215:1091–100. doi: 10.1084/jem.20171068
179. Blank CU, Haining WN, Held W, Hogan PG, Kallies A, Lugli E, et al. Defining 'T Cell Exhaustion'. Nat Rev Immunol (2019) 19:665–74. doi: 10.1038/s41577-019-0221-9
180. Sadelain M, Riviere I, Riddell S. Therapeutic T Cell Engineering. Nature (2017) 545:423–31. doi: 10.1038/nature22395
181. Solinas C, De Silva P, Bron D, Willard-Gallo K, Sangiolo D. Significance of TIM3 Expression in Cancer: From Biology to the Clinic. Semin Oncol (2019) 46:372–9. doi: 10.1053/j.seminoncol.2019.08.005
182. Hong M, Clubb JD, Chen YY. Engineering CAR-T Cells for Next-Generation Cancer Therapy. Cancer Cell (2020) 38:473–88. doi: 10.1016/j.ccell.2020.07.005
183. Fraietta JA, Lacey SF, Orlando EJ, Pruteanu-Malinici I, Gohil M, Lundh S, et al. Determinants of Response and Resistance to CD19 Chimeric Antigen Receptor (CAR) T Cell Therapy of Chronic Lymphocytic Leukemia. Nat Med (2018) 24:563–71. doi: 10.1038/s41591-018-0010-1
184. Cherkassky L, Morello A, Villena-Vargas J, Feng Y, Dimitrov DS, Jones DR, et al. Human CAR T Cells With Cell-Intrinsic PD-1 Checkpoint Blockade Resist Tumor-Mediated Inhibition. J Clin Invest (2016) 126:3130–44. doi: 10.1172/JCI83092
185. Sanghera C, Sanghera R. Immunotherapy - Strategies for Expanding Its Role in the Treatment of All Major Tumor Sites. Cureus (2019) 11:e5938. doi: 10.7759/cureus.5938
186. Lynn RC, Weber EW, Sotillo E, Gennert D, Xu P, Good Z, et al. C-Jun Overexpression in CAR T Cells Induces Exhaustion Resistance. Nature (2019) 576:293–300. doi: 10.1038/s41586-019-1805-z
187. Morgan MA, Büning H, Sauer M, Schambach A. Use of Cell and Genome Modification Technologies to Generate Improved "Off-The-Shelf" CAR T and CAR NK Cells. Front Immunol (2020) 11:1965. doi: 10.3389/fimmu.2020.01965
Keywords: chimeric antigen receptor, T cell, immunotherapy, lung cancer, engineering strategy
Citation: Xiao B-F, Zhang J-T, Zhu Y-G, Cui X-R, Lu Z-M, Yu B-T and Wu N (2021) Chimeric Antigen Receptor T-Cell Therapy in Lung Cancer: Potential and Challenges. Front. Immunol. 12:782775. doi: 10.3389/fimmu.2021.782775
Received: 24 September 2021; Accepted: 13 October 2021; Published: 01 November 2021.
Reviewed by:
Copyright © 2021 Xiao, Zhang, Zhu, Cui, Lu, Yu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY) . The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ben-Tong Yu, [email protected] ; Nan Wu, [email protected]
† These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Lung Cancer Research Results and Study Updates
See Advances in Lung Cancer Research for an overview of recent findings and progress, plus ongoing projects supported by NCI.
The results of the clinical trial that led to FDA’s 2023 approval of repotrectinib (Augtyro) for lung cancers with ROS1 fusions have been published. The drug shrank tumors in 80% of people receiving the drug as an initial treatment.
Tarlatamab, a new type of targeted immunotherapy, shrank small cell lung cancer (SCLC) tumors in more than 30% of participants in an early-stage clinical trial. Participants had SCLC that had progressed after previous treatments with other drugs.
For people with lung cancer and medullary thyroid cancer whose tumors have changes in the RET gene, selpercatinib improved progression-free survival compared with other common treatments, according to new clinical trial results.
In the ADAURA clinical trial, people with early-stage lung cancer treated with osimertinib (Tagrisso) after surgery lived longer than people treated with a placebo after surgery. Despite some criticisms about its design, the trial is expected to change patient care.
For certain people with early-stage non-small cell lung cancer, sublobar surgery to remove only a piece of the affected lung lobe is as effective as surgery to remove the whole lobe, new research shows.
Pragmatica-Lung is a clinical trial for people with non-small cell lung cancer that has spread beyond the lungs (stage 4 cancer). The trial will help confirm if the combination of pembrolizumab and ramucirumab helps people with advanced lung cancer live longer.
On August 11, the Food and Drug Administration (FDA) gave accelerated approval to trastuzumab deruxtecan (Enhertu) for adults with non-small cell lung cancer (NSCLC) that has a specific mutation in the HER2 gene. Around 3% of people with NSCLC have this kind of HER2 mutation.
Giving people with early-stage lung cancer the immunotherapy drug nivolumab (Opdivo) and chemotherapy before surgery can substantially delay the progression or return of their cancer, a large clinical trial found.
Atezolizumab (Tecentriq) is now the first immunotherapy approved by FDA for use as an additional, or adjuvant, treatment for some patients with non-small cell lung cancer. The approval was based on results of a clinical trial called IMpower010.
Quitting smoking after a diagnosis of early-stage lung cancer may help people live longer, a new study finds. The study, which included more than 500 patients, also found that quitting smoking delayed the cancer from returning or getting worse.
NCI scientists and their international collaborators have found that the majority of lung cancers in never smokers arise when mutations caused by natural processes in the body accumulate. They also identified three subtypes of lung cancer these individuals.
FDA has approved the first KRAS-blocking drug, sotorasib (Lumakras). The approval, which covers the use of sotorasib to treat some patients with advanced lung cancer, sets the stage for other KRAS inhibitors already in development, researchers said.
Combining the chemotherapy drug topotecan and the investigational drug berzosertib shrank tumors in some patients with small cell lung cancer, results from an NCI-supported phase 1 clinical trial show. Two phase 2 trials of the combination are planned.
Mortality rates from the most common lung cancer, non-small cell lung cancer (NSCLC), have fallen sharply in the United States in recent years, due primarily to recent advances in treatment, an NCI study shows.
In a study of more than 50,000 veterans with lung cancer, those with mental illness who received mental health treatment—including for substance use—lived substantially longer than those who didn’t participate in such programs.
FDA has granted accelerated approval for selpercatinib (Retevmo) to treat certain patients with thyroid cancer or non-small cell lung cancer whose tumors have RET gene alterations. The drug, which works by blocking the activity of RET proteins, was approved based on the results of the LIBRETTO-001 trial.
Osimertinib (Tagrisso) improves survival in people with non-small cell lung cancer with EGFR mutations, updated clinical trial results show. People treated with osimertinib lived longer than those treated with earlier-generation EGFR-targeted drugs.
A large clinical trial showed that adding the immunotherapy drug durvalumab (Imfinzi) to standard chemotherapy can prolong survival in some people with previously untreated advanced small cell lung cancer.
The investigational drug selpercatinib may benefit patients with lung cancer whose tumors have alterations in the RET gene, including fusions with other genes, according to results from a small clinical trial.
FDA has approved entrectinib (Rozlytrek) for the treatment of children and adults with tumors bearing an NTRK gene fusion. The approval also covers adults with non-small cell lung cancer harboring a ROS1 gene fusion.
Clinical recommendations on who should be screened for lung cancer may need to be reviewed when it comes to African Americans who smoke, findings from a new study suggest.
Use of a multipronged approach within hospitals, including community centers, not only eliminated treatment disparities among black and white patients with early-stage lung cancer, it also improved treatment rates for all patients, results from a new study show.
In everyday medical care, there may be more complications from invasive diagnostic procedures performed after lung cancer screening than has been reported in large studies.
The Lung Cancer Master Protocol, or Lung-MAP, is a precision medicine research study for people with advanced non-small cell lung cancer that has continued to grow after treatment. Patients are assigned to different study drug combinations based on the results of genomic profiling of their tumors.
On December 6, 2018, the Food and Drug Administration (FDA) approved atezolizumab (Tecentriq) in combination with a standard three-drug regimen as an initial treatment for advanced lung cancer that does not have EGFR or ALK mutations.
A new study has identified a potential biomarker of early-stage non–small cell lung cancer (NSCLC). The biomarker, the study’s leaders said, could help diagnose precancerous lung growths and early-stage lung cancers noninvasively and distinguish them from noncancerous growths.
Results from two large clinical trials should cement the value of the drugs brigatinib (Alunbrig) and durvalumab (Imfinzi) in treating non-small cell lung cancer (NSCLC). The trial results, several experts said, confirm that the drugs can improve the outcomes of patients with advanced NSCLC.
Cancer researchers have trained a computer program to scan images of tissue samples to differentiate normal lung tissue from the two most common forms of lung cancer. The program also learned to detect cancer-related genetic mutations in the samples.
A collection of material about the ALCHEMIST lung cancer trials that will examine tumor tissue from patients with certain types of early-stage, completely resected non-small cell lung cancer for gene mutations in the EGFR and ALK genes, and assign patients with these gene mutations to treatment trials testing post-surgical use of drugs targeted against these mutations.

Lung Cancer Cells Switch Oncogenic Drivers
Mouse models mimicking the transition from a common form of lung cancer to a more aggressive one may help scientists develop future strategies to prevent this transformation..

Alejandra Manjarrez is a freelance science journalist who contributes to The Scientist . She has a PhD in systems biology from ETH Zurich and a master’s in molecular biology from Utrecht University.
View full profile.
Learn about our editorial policies.
ABOVE: Researchers developed experimental models that mimic the histological transformation from lung adenocarcinoma to small cell lung cancer in mice. Eric Gardner, Weill Cornell Medicine
L ung adenocarcinoma (LUAD), the most common form of lung cancer, is already a challenging disease. It is currently the leading cause of cancer death in the United States. 1 In response to therapies that inhibit their genetic drivers, LUAD cells may undergo transformation as a mechanism of resistance and begin to exhibit traits of a more aggressive and difficult-to-treat cancer type known as small cell lung cancer (SCLC).
To gain deeper insight into the mechanisms underlying this problematic transition, Eric Gardner , a lung cancer biologist at Weill Cornell Medicine, and his colleagues developed mouse models that recapitulated the process. The researchers found that the lung cell type that gives rise to LUAD is typically unaffected by mutations in one of the oncogenes driving SCLC. However, when the team inhibited an oncogenic driver for LUAD and deleted specific tumor suppressor genes, a subset of LUAD cells converted into a stem-like state. Under these conditions, the cells were plastic enough to respond to the cancer gene driving the SCLC transformation, the team reported in Science . 2
Gardner’s team first focused their efforts on the challenging task of creating mouse strains that could recapitulate the complex transformation in a simplified model. “It took three years to make these mice, and then it took another two to three years to actually model this process,” said Gardner.
The design of these genetically engineered mice was inspired by other mouse models for different lung cancer types. “I didn’t really develop anything that new so much as I took a little bit of this, a little bit of that, and brought it together,” he explained. The resulting mouse strains had mechanisms to turn on the expression of cancer genes involved in either LUAD or SCLC in specific cell types, for example.
Researchers also labelled the mouse tumor cells with a fluorescent protein to track their course and used single-cell RNA sequencing to characterize gene expression throughout the transformation.

The models revealed that lung alveolar epithelial cells that give rise to LUAD are sensitive to mutations in the oncogene coding for the epidermal growth factor receptor (EGFR), but are resistant to transformation driven by overexpression of Myc , one of the cancer genes leading to SCLC. Conversely, pulmonary neuroendocrine cells, the cells of origin for SCLC, are responsive to Myc overexpression, but not to EGFR mutations.
Gardner and his colleagues explored the mechanisms that rendered alveolar epithelial cells susceptible to the oncogenic effects of Myc . The researchers discovered that by deleting specific tumor suppressor genes—some previously linked to SCLC—and blocking EGFR function, a subset of the alveolar epithelial cells transformed into stem-like cells that are sensitive to both EGFR and Myc . This plasticity allowed these cells to transform into Myc -driven SCLC neuroendocrine tumor cells. “This represents . . . a fundamental switch in the oncogenic driver program,” said Gardner.
The advantage of this system is that it enables researchers to turn on and off the expression of lineage-specific oncogenic drivers, according to Hideo Watanabe , a genomic scientist studying lung cancer lineages at the Icahn School of Medicine at Mount Sinai who did not participate in this study. “It’s a very elegant [and] laborious way to look at this [transition],” he said.
Watanabe added that these mouse models may help scientists identify potential biomarkers activated during the transition from LUAD to SCLC, which could be monitored in patients to guide treatment decisions. However, he cautioned that researchers still need to determine how accurately the molecular processes in the model reflect LUAD to SCLC transformation in human lung cancer. Moreover, he noted, this might represent just one of many transition mechanisms, given the heterogeneity of lung cancer.
Watanabe believes that there's a long way to go before these findings have direct clinical implications. Yet, he added that the study offers foundational information that may help scientists develop future strategies for preventing this transformation in patients.

- Myers DJ, Wallen JM. Lung adenocarcinoma . StatPearls [Internet]. 2023.
- Gardner E, et al. Lineage-specific intolerance to oncogenic drivers restricts histological transformation . Science . 2024;383(6683):eadj1415.
Related cancer cell biology Research Resources

Optimizing Organoid Culture for Development and Disease Modeling


Viewing the Glioblastoma Tumor Microenvironment at Single Cell Resolution

- Open access
- Published: 15 May 2023
Decoding the key compounds and mechanism of Shashen Maidong decoction in the treatment of lung cancer
- Jieqi Cai 1 , 2 na1 ,
- Yupeng Chen 1 , 2 na1 ,
- Kexin Wang 3 na1 ,
- Yi Li 1 , 2 ,
- Jie Wu 1 , 2 ,
- Hailang Yu 1 , 2 ,
- Qingping Li 4 ,
- Wei Meng 1 , 2 ,
- Handuo Wang 1 , 2 ,
- Aiping Lu 6 ,
- Mianbo Huang 7 ,
- Genxia Wei 8 &
- Daogang Guan 1 , 2
BMC Complementary Medicine and Therapies volume 23 , Article number: 158 ( 2023 ) Cite this article
1470 Accesses
2 Citations
Metrics details
Lung cancer is a malignant tumour with the fastest increase in morbidity and mortality around the world. The clinical treatments available have significant side effects, thus it is desirable to identify alternative modalities to treat lung cancer. Shashen Maidong decoction (SMD) is a commonly used traditional Chinese medicine (TCM) formula for treating lung cancer in the clinic. While the key functional components (KFC) and the underlying mechanisms of SMD treating lung cancer are still unclear.
We propose a new integrated pharmacology model, which combines a novel node-importance calculation method and the contribution decision rate (CDR) model, to identify the KFC of SMD and to deduce their mechanisms in the treatment of lung cancer.
The enriched effective Gene Ontology (GO) terms selected from our proposed node importance detection method could cover 97.66% of enriched GO terms of reference targets. After calculating CDR of active components in key functional network, the first 82 components covered 90.25% of the network information, which were defined as KFC. 82 KFC were subjected to functional analysis and experimental validation. 5–40 μM protocatechuic acid, 100–400 μM paeonol or caffeic acid exerted significant inhibitory activity on the proliferation of A549 cells. The results show that KFC play an important therapeutic role in the treatment of lung cancer by targeting Ras, AKT, IKK, Raf1, MEK, and NF-κB in the PI3K-Akt, MAPK, SCLC, and NSCLC signaling pathways active in lung cancer.
Conclusions
This study provides a methodological reference for the optimization and secondary development of TCM formulas. The strategy proposed in this study can be used to identify key compounds in the complex network and provides an operable test range for subsequent experimental verification, which greatly reduces the experimental workload.
Peer Review reports
Introduction
Lung cancer is the most common primary malignancy and ranks first in the incidence and mortality of malignant tumors in the world. The incidence of lung cancer has reached 11.6% of total cancer cases, and mortality is 18.4% of total cancer deaths [ 1 ]. At present, lung cancer treatments mainly include surgery, radiation, and chemotherapy. These treatments can prevent the malignant development of tumor cells and improve survival time. However, most treatments present obvious side effects in clinical use, and can cause serious or general damage to specific or all tissues, including functional damage to the digestive system and urinary system [ 2 ]. In addition to the above treatments, some targeted drugs are often used in the clinical treatment of lung cancer, including gefitinib [ 3 ], bevacizumab [ 4 ], and osimertinib [ 5 ], but with these drugs it is difficult to control toxicity and side effects. Thus, it is desirable to improve the treatment for lung cancer.
Many clinical studies have shown that anti-lung cancer treatment combined with Traditional Chinese Medicine (TCM) can reduce the side effects of radiotherapy and chemotherapy, improve therapeutic effects, and reduce complications. Meanwhile, TCM can prolong the life of lung cancer patients and improve their survival quality of life [ 6 ]. Currently, some studies have shown that TCM formulas could effectively treat lung cancer. For example, a well-known Yangyinwenyang formula was used to induce lung cancer cell apoptosis in vitro [ 7 ]. Jinfukang’s formula could inhibit lung cancer cell proliferation by promoting cell apoptosis [ 8 ]. Kangliuzengxiao decoction is successful for the prevention the development of tumors and improving major clinical symptoms [ 9 ]. Shashen Maidong Decoction (SMD) can alleviate symptoms of patients and can reduce the inflammatory response [ 10 ]. Among these formulas, SMD is widely used in the clinical treatment of lung cancer. As reports, based on the cancer toxicity theory, the application of SMD in treating lung cancer cachexia has definite therapeutic effects and important clinical values [ 10 ]. The carcinoembryonic antigen (CEA) and carbohydrate antigen 153 (CA 153) level was significantly reduced in serum of two patient groups [ 11 ]. In addition, SMD is proved to have obvious inhibition on the growth of A549 cells. In the result of MTT assay, the growth of A549 cells was inhibited obviously in the SMD-containing serum [ 12 , 13 ]. A549 cells cultured in the SMD-containing serum showed higher E-cadherin protein expression, lower Snail protein expression ( P < 0.05) [ 12 ], higher Smad7 protein expression and lower TGF-β1 protein expression [ 13 ].
SMD is composed of 7 herbs, Glehnia littoralis (A.Gray) F.Schmidt ex Miq. ( G. littoralis , Beishashen, 9 g), Ophiopogon japonicus (Thunb.) Ker Gawl. ( O. japonicus , Maidong, 9 g), Polygonatum odoratum (Mill.) Druce ( P. odoratum, Yuzhu, 6 g), Trichosanthes kirilowii Maxim. ( T. kirilowii , Tianhuafen, 4.5 g), Lablab purpureus subsp. purpureus ( L. purpureus, Baibiandou, 4.5 g), Morus alba L. ( M. alba, Sangye, 4.5 g), and Glycyrrhiza uralensis Fisch. ex DC. ( G. uralensis , Gancao, 3 g). O. japonicus and G. littoralis are the main drugs of SMD, and may be responsible for the therapeutic effects of SMD in lung cancer. Modern pharmacological research indicates that the extract of O. japonicus has an obvious inhibitory effect on the occurrence and development of lung cancer, and it can induce autophagy of A549 lung cancer cells [ 14 ]. G. littoralis exerts anticancer activity and induces cycle arrest of A549 cells [ 15 ]. In clinical research, SMD combined with hormones or antibiotics is a remarkable treatment for radiation pneumonia [ 16 ]. Furthermore, chemotherapy combined with modified SMD has been shown to be highly effective in the cure of mid- and late-stage non-small cell lung cancer (NSCLC) [ 17 ]. But there is still a lack reports characterizing key functional components (KFC) and the potential therapeutic mechanism of SMD on lung cancer at a system level.
Due to the “multi-components-multi-targets” of the formula, it is difficult to explore the potential associations among TCM components, target genes, and diseases using traditional experiments. Based on the theory of systems biology, integrated pharmacology can clarify the synergistic mechanism of the components-targets and of the targets-pathogenic network [ 18 ]. It is important to study the mechanisms underlying the therapeutic activity of TCM. The “multi-components-multi-targets” characteristic of TCM formulas determine that it could form complex regulation relations between components and targets in the treatment process. Among these complex relationships, some relationships have positive effects on treatment, while others have antagonistic or toxic effects. The aim of formula optimization is a process that finds components with synergistic effects and removes components with antagonistic or toxic effects [ 19 ]. Optimization of the formula would be beneficial for the secondary development of TCM.
In this study (Fig. 1 ), the chemicals of SMD were collected from published databases, then the active components were screened from all components based on Lipinski’s rule, oral bioavailability (OB) value, and gastrointestinal (GI) absorption. The targets of active components were predicted using on-line tools, the active components and their targets were used to construct the components-targets (CT) network. Meanwhile, pathogenic genes associated with lung cancer were collected from DisGeNET [ 20 ] to construct a network of weighted pathogenic genes. The CT network and the weighted pathogenic gene network were then integrated to construct the CTP network. Finally, a node importance calculation method was designed to select the key functional network and effective proteins. The contribution decision rate (CDR) model was developed to capture KFC. Finally, KFC and their targets were used to infer the potential mechanism of SMD in the treatment of lung cancer.
The work scheme of our proposed integrated pharmacology approach
Materials and methods
Identification of the components of shashen maidong decoction and chemical analysis.
The component structures of SMD in MOL2 format were downloaded from the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) [ 21 ], the Encyclopedia of Traditional Chinese Medicine (ETCM) [ 22 ], and Symptom Mapping (SymMap) [ 23 ]. The components in different formats were converted into a unified SMILES format by Open Babel 2.4.1 [ 24 ]. We searched for experimentally validated high concentrations and biological activities of chemical components of SMD from the literature to obtain a comprehensive list of active components.
Selection of potential active components based on published ADMET models
In general, effective drugs have good absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties. Therefore, in the process of screening possible active components of SMD, we considered some ADMET properties, including Lipinski’s rule [ 25 ], GI absorption value [ 26 ], and OB [ 21 ].
The detailed criteria of Lipinski’s Rule are as follows: there are no more than 5 H-bond donors, less than 10 H-bond acceptors, the molecular weight (MW) is not greater than 500, the calculated Log P is not greater than 5 and the number of rotatable bonds in compounds is less than 10. Ingredients that meet these standards have better drug-like properties.
OB (%F) represents the percentage of oral dose of the chemical components of botanical drugs that are released into the systemic circulation. The chemical components with OB ≥ 30% are well absorbed in human body, so these chemical components were selected for further study.
GI absorption is a pharmacokinetic property, and it is vital to estimate the GI absorption value at each stage of the drug discovery process. GI absorption values of all components in SMD were obtained from SwissADME and chemical components with high GI values were retained for further research.
Predicting the molecular targets of the active components
To obtain the molecular targets of the active components in SMD, we used SwissTargetPrediction [ 27 ], HitPick [ 28 ], and the Similarity Ensemble Approach (SEA) [ 29 ] to predict the potential molecular targets.
Constructing the weighted pathogenic genes network of lung cancer
In the DisGeNET database [ 20 ], we identified a total of 27 lung cancer-related international classification of diseases (ICD, version 10) codes by querying “lung cancer” as the key word. We then used these classification IDs to query related genes; a total of 2973 pathogenic genes were obtained.
Definition of protein–protein interaction data
We submitted 2973 pathogenic genes and 1221 predicted targets of active components to the STRING [ 30 ] database and chose “Homo sapiens” as the species parameter to obtain the protein–protein interaction (PPI) network.
Construction of component-target network
All SMD chemicals collected in the database were screened with ADMET to obtain active components, and the targets of these active components were predicted by Hitpick, SwissTargetPrediction, and SEA. Active components and their predicted targets were used to construct components-targets (CT) network using Cytoscape [ 31 ].
Screening of key functional networks
We constructed a four-layer component-target-gene-disease network by combining CT, PPI network, and pathogenic gene network, and we determined and verified the key component-target-pathogenic gene associations in the network with our newly designed node-importance calculation method. The detailed algorithm was described in the Supporting information .
Prediction of key functional components
The Contribution Decision Rate (CDR) model, which was employed and modified based on knapsack algorithm, was used to determine the KFC in the network: Assuming there is a function that can solve the total value with two dependent variables n and C, the detailed algorithm was described in the Supporting information .
GO and KEGG analysis
R package clusterProfiler [ 32 ] was used to perform the Kyoto Encyclopedia of Genes and Genomes (KEGG) [ 33 , 34 , 35 ] and Gene Ontology (GO) enrichment analysis, and P -value < 0.05 was set as significant level. After that, we used ggplot2 [ 36 ] to create the graph of enrichment results. Then, some specific and enriched pathways were merged and visualized to speculate on the potential mechanism of SMD on lung cancer.
Cell culture and treatment
Human lung cancer A549 cells were obtained from GuangZhou Jennio Biotech Co., Ltd. Fetal bovine serum (FBS) and Dulbecco’s modified Eagle’s medium (DMEM) were purchased from Gibco (Grand Island, USA). A549 cells were cultured in DMEM, containing 10% FBS, 100 units/mL of penicillin, and 100 μg/mL of streptomycin at 37℃ under 5% CO2. Protocatechuic acid (≥ 99.5% purity by HPLC), paeonol (≥ 99% purity by HPLC), and caffeic acid (≥ 98% purity by HPLC), which purchased form Jiangsu Yongjian Pharmaceutical Technology Co.,Ltd, were dissolved in DMSO. When the cells reached 80% confluence, they were treated with various concentrations of protocatechuic acid (1, 5, 10, 20, 40 μM), paeonol (25, 50, 100, 200, 400 μM) or caffeic acid (25, 50, 100, 200, 400 μM) for 24 h, respectively. The control group was not treated with drug components.
The concentration of our experimental components was determined referring to public reports. As reported, 80 or 100 μg/mL Paeonol caused a significant effect on cell viability of A549 cells compared to 0 μg/mL Paeonol [ 37 , 38 ]. Caffeic acid with a concentration higher than 100 μM has significant cytotoxic effect on A549 cells [ 39 ]. Protocatechuic acid at 2–8 micromol/L significantly inhibited A549 cell adhesion ( P < 0.05) [ 40 ].
Cell viability assay
A549 cells (1 × 105 cells/well) were planted in 96-well plates. After a 24-h culture, the cells were treated with different concentrations of protocatechuic acid (1, 5, 10, 20, 40 μM), paeonol (25, 50, 100, 200, 400 μM), or caffeic acid (25, 50, 100, 200, 400 μM) for 24 h, respectively. Subsequently, each well was added with 10 μL of MTT solution (5 mg/ml). After 4 h, the supernatant was removed and 100 μL of DMSO was supplied into each well to dissolve the MTT formazan product. The absorbance was measured at 570 nm using the Infinite M200 PRO plate reader (Tecan, Switzerland).
Collection of chemical components and screening of active components in SMD
A total of 523 types of chemical components of SMD were retrieved from TCMSP, ETCM, and SymMap databases (Additional Table 1 ). Among these components, some had higher concentration than expected in SMD by UPLC-MS/MS method (Table 1 ) [ 41 ], such as rutin, liquiritin, psoralen, xanthotoxin, bergapten, monoammonium glycyrhizinate, ophiopogonin D, methylophiopogonanone A, and methylophiopogonanone B. Previous reports confirmed that high-concentration components of SMD may play important roles in the treatment of lung cancer. By combining the experimental validated high-concentration components (Table 1 ) [ 41 ], and ADMET model predicted components, 284 active components were figured out for subsequent analysis (Additional Table 2 , Fig. 2 ).
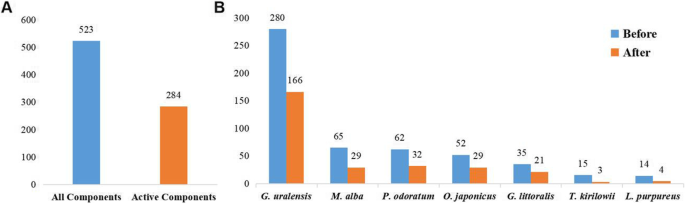
Distribution of all components and active components. A All components and active components of SMD. B All components and active components of each herb in SMD
Among all active components, 14 components shared two or multiple herbs in SMD (Fig. 3 ), quercetin, isoquercitrin, rutin, scopoletol, glucuronic acid, 2-pentylfuran, beta-carotene, gynesine, arachic acid, 2-heptanone, linoleic acid, palmitic acid, oleic acid, and octadiene, have been shown to have clear anticancer effects. For example, quercetin has definite anticancer activity and can inhibit a variety of carcinogenic signaling pathways [ 42 ]. Rutin promotes the apoptosis of TNF-α-induced A549 human lung cancer cells [ 43 ]. Scopoletol can play an anticancer role by triggering apoptosis, blocking the cell cycle, inhibiting cell invasion, and regulating the PI3K/AKT signaling pathway [ 44 ]. Quercetin, isoquercitrin, rutin, and scopoletol are also present in G. littoralis , M. alba and G. uralensis , which may be of great importance in the therapeutic mechanism of SMD on lung cancer. These results indicate that shared components may play important therapeutic roles in lung cancer.
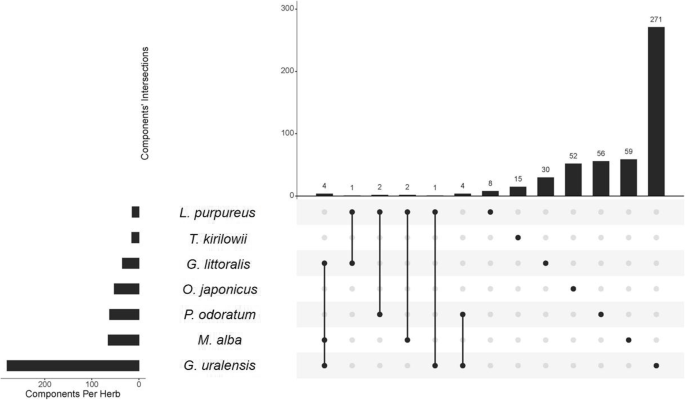
Common and specific active components in different herbs of SMD. The histogram shows the number of components common to different herbs, and the dark dots represent the original herb of the intersection
Predicting targets of active components in SMD
The 1221 targets of 284 active components were predicted by SwissTargetPrediction, HITPICK, and SEA (Additional Table 3 ). The active components and their targets were used to construct the CT network. This network contains 1505 nodes and 11,505 interactions (Fig. 4 ). We analyzed the degree of each component and each target in this network and found that the average degree of the components was 40.51 and the average degree of the targets was 9.42. Ten components with the highest degree were vanillic acid, ferulic acid, dibenzoylmethane, butyl benzoate, lauric acid, nicotinic acid, dibutyl phthalate, salicylic acid, eugenol, n-cis-feruloyltyramine. Most have been reported to be associated with cancer therapy, vanillic acid has antioxidant activity in scavenging free radicals, and thus, it has an effective preventive effect on lung cancer [ 45 ]. Lauric acid can be used as a carrier of targeted drugs due to its specific accumulation in lung cancer tissues [ 46 ]. The ten targets with the highest degree of the components were MAPT, ESR1, TDP1, ESR2, CYP1B1, ABCG2, ODC1, PTPN1, CYP19A1, and TYR. Most of these genes are reported to be related to the pathogenesis of lung cancer. MAPT has been shown to induce lung cancer cells to gain taxol resistance by activating the PI3K/Akt signaling pathway [ 47 ]. ESR1 is a central gene of lung cancer and promotes the occurrence of lung cancer by regulating the p53 signaling pathway and the cell surface receptor signaling pathway [ 48 ]. Overexpression of TDP1 is closely related to tumorigenesis, and is a crucial target for tumor treatment, the TDP1 inhibitor can significantly increase the antitumor effect of drugs [ 49 ]. These analyses demonstrate that one component can regulate multiple targets and in turn, one target is regulated by multiple components, which reflects the characteristics of the “multicomponents-multitargets” theory in treating complex diseases of TCM.
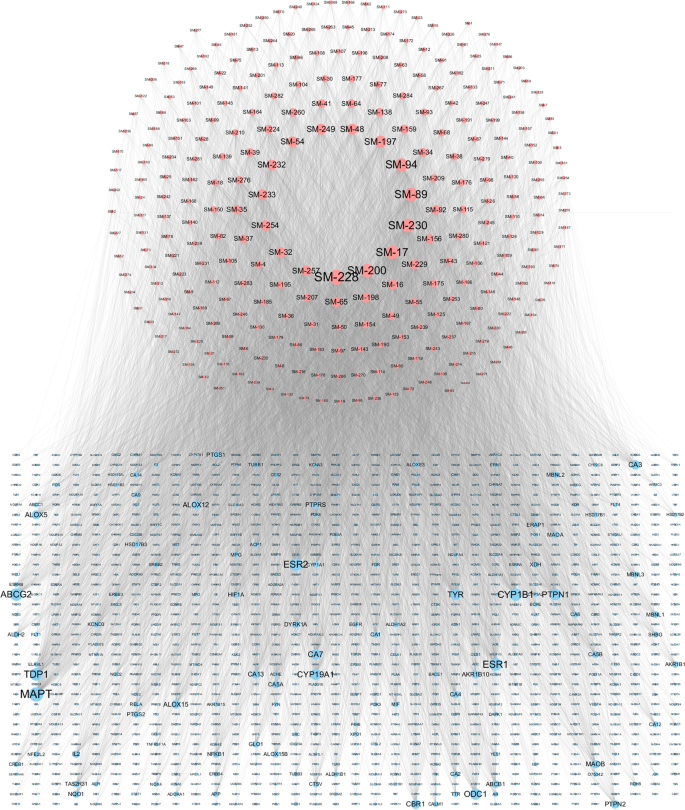
Pharmaceutical component-target interaction network. The red nodes represent pharmaceutical components, and the blue nodes refer to the targeted genes. The size of the nodes represents their degree
Construction of the pathogenic genes weighted network
The pathogenic gene data set was retrieved and downloaded from the DisGeNET database and 2973 pathogenic genes with the number of supporting publications greater than or equal to the mean value were retained (Additional Table 4 ). The number of supporting publications reflects the correlation of genes associated with lung cancer. A higher number of supporting publications suggests that genes have more associations with lung cancer. After counting the number of supporting publications for 2973 pathogenic genes (Fig. 5 A), we found that more than half of these pathogenic genes have only one supporting reference and 33 pathogenic genes with more than 40 supporting references. The top ten genes with the highest number of supporting publications are EGFR, TP53, KRAS, ALK, GSTM1, CDKN2A, CYP1A1, ERBB2, BCL2, and MET. The epidermal growth factor receptor (EGFR) is a transmembrane glycoprotein of the ErbB family of tyrosine kinase receptors, and activated mutations of EGFR are a remarkable feature of lung cancer [ 50 ]. EGFR-activating mutations are highly sensitive to tyrosine kinase inhibitor gefitinib, so gefitinib is commonly used in clinical targeted treatment of NSCLC [ 51 ]. TP53-induced glycolytic phosphatase (TIGAR) is a key regulator of glycolysis and apoptosis, which can protect cells from oxidative stress-induced apoptosis and provide the necessary conditions for the survival of cancer cells [ 52 ]. Mutations exist widely in many types of lung cancer [ 53 , 54 ]. KRAS is a potential oncogene and has been reported to have a high mutation rate, which makes cancer cells escape apoptosis-induced cell death [ 55 ]. The rearrangement of anaplastic lymphoma kinase (ALK) plays an important role in promoting the occurrence and development of lung cancer [ 56 ], thus it becomes an important clinical targeted treatment of ALK-positive NSCLC [ 57 ]. To explore whether it is reliable to measure the importance of gene function by the number of relevant supporting publications, we constructed a KEGG and GO analysis of pathogenic genes of lung cancer (Fig. 5 B). The results showed that there is a positive correlation between supporting publications and the functional pathways involved by these genes, as well as GO terms. Genes with more supporting publications are associated with a highest number of pathways involved.
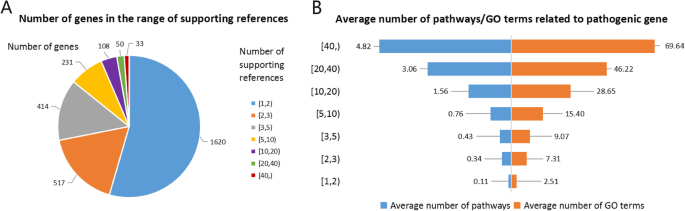
The number of supporting publications and involved pathways of pathogenic genes in lung cancer. A The number and distribution of supporting publications; B Average number of pathways and GO terms related to pathogenic gene in a distinct interval of supporting publications
Key functional network selection and validation
In formulas that treat complex diseases, some components play major therapeutic roles, some play auxiliary roles, and others play antagonistic roles. The group of components with major therapeutic roles is usually considered KFC. The KFC and their targets form key functional network, embedded within the complex CT network. How to detect the key functional network and KFC is the basis for optimizing TCM formulations. Additionally, the process of drug treatment of diseases is a continuous process of drug interventions through protein–protein interactions. Based on this information, we integrated the CT network and pathogenic gene weighted network to construct the comprehensive CTP network (Additional Fig. 1 ). Then we designed a new node importance calculation method to capture the key functional network. The nodes larger than the median important values of all nodes in the network were retained, and these nodes and their interactions were defined as the key functional network.
To test the reliability of the node importance calculation method, we first performed GO enrichment analysis on targeted genes and pathogenic genes of lung cancer and considered the intersection of GO terms as the effective GO terms to serve as a reference for further comparison. Comparing our proposed node importance detection method with the other traditional methods (Fig. 6 A), such as Radiality, Closeness to center, Degree, Neighborhood Connectivity Clustering coefficient and Average Shortest Path length, we found that our method covered up to 97.66% of effective GO terms, which is higher than Radiality 96.02%, Degree 95.74%, Neighborhood Connectivity 86.66%, and the Clustering Coefficient 77.54%. Figure 6 B shows that our model also covered the most pathways, thus it indicated that the key functional network detection model we designed could retain the key intervention information.
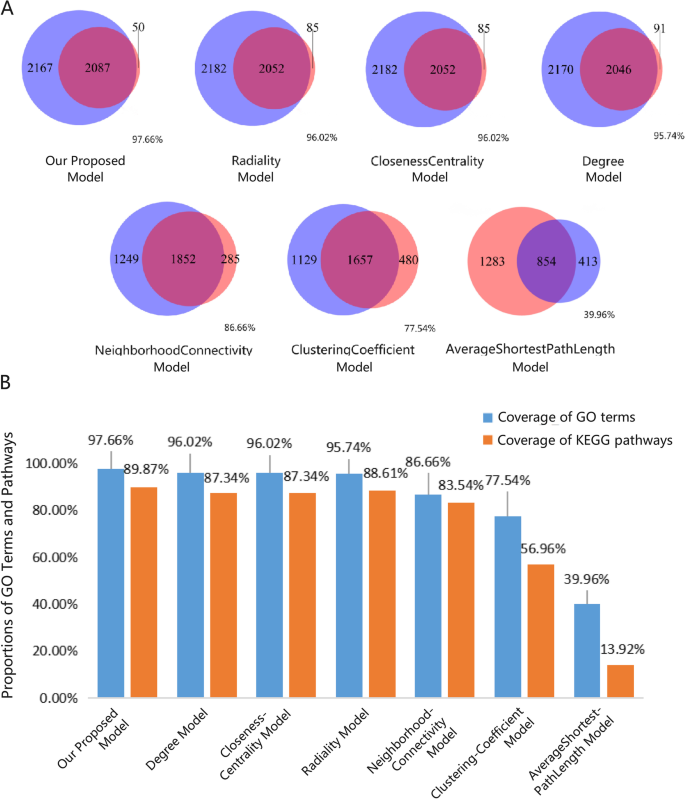
Validation of key functional networks. A The Venn diagrams display the number of elements of the seven models that overlap with effective GO terms. The red cycle represents the effective GO terms, and the blue cycle represents the GO terms predicted by different models. B Comparison of our proposed models with other traditional models on the coverage of enriched GO terms and pathways
Find key functional components
After extracting the key functional network, we used the CDR model to deduce which components could retain the key functional network information to the maximum extent. Finally, we obtained 82 components that defined the KFC (Additional Table 5 , Fig. 7 ). In the KFC, the CDR of the first 8 components reached 50% target coverage, and the 82 components reached 90% target coverage. Among these components, vanillic acid has been reported to have antioxidant activity in scavenging free radicals and had a significant and effective preventative role in B(a)P-induced lung cancer [ 45 , 46 ]. Lauric acid is a good carrier of targeted drugs for lung cancer [ 46 ] and can also inhibit the expression of carcinogenic miRNA and significantly up-regulate the expression of some cancer-inhibiting miRNA in KB cells and HepG2 cells [ 58 ]. Salicylic acid can maintain the stability of the genome and plays a key role in reducing the risk of cancer. These findings indicate that the lack of salicylic acid will lead to the delay of DNA excision and repair mechanisms, the accumulation of single-strand and double-strand breaks, cell cycle arrest, damage from apoptosis, and will increase the susceptibility to cancer development [ 59 ].
The CDR model of active components in lung cancer
Effects of key functional components on the viability of A549 cells
Simple random sampling is a method of probability sampling based on chance events, and it can mitigate selection bias [ 60 ]. It’s the incorporation of randomization that provides unpredictability in treatment assignments. Based on this selective strategy, protocatechuic acid (SM-32), paeonol (SM-257), and caffeic acid (SM-229) in 82 KFC were selected to determine the effects on the viability of A549 cells using the MTT assay. Protocatechuic acid (SM-32) originates from G. uralensis . Paeonol (SM-257) originates from M. alba . Caffeic acid (SM-229) originates from G. littoralis . After 24 h of incubation, the cell viabilities of A549 cells were 95.07 ± 8.30%, 85.31 ± 6.08%, 60.65 ± 4.01%, 48.25 ± 10.39%, and 39.12 ± 5.88% after exposure to protocatechuic acid at concentrations of 1, 5, 10, 20, and 40 μM, respectively (Fig. 8 A). After exposure to paeonol at concentrations of 25, 50, 100, 200 and 400 μM, the cell viability was 97.34 ± 6.58%, 94.33 ± 9.72%, 89.77 ± 6.94%, 75.19 ± 13.70% and 66.79 ± 11.76% (Fig. 8 B). When cells were treated with 25, 50, 100, 200, and 400 μM caffeic acid, the cell viabilities were 95.60 ± 7.97%, 93.90 ± 7.26%, 90.27 ± 6.18%, 79.91 ± 5.54%, and 60.89 ± 5.39% (Fig. 8 C). The results show that 5–40 μM protocatechuic acid, 100–400 μM paeonol or caffeic acid exerted significant inhibitory activity on the proliferation of A549 cells.
Inhibitory Effects of protocatechuic Acid A , paeonol B and caffeic Acid C on the proliferation of A549 cells at 24 h. Data are represented as mean ± SEM ( n = 6). * P < 0.05, ** P < 0.01, *** P < 0.001 versus the control group. The 2D structures are obtained from PubChem
Possible therapeutic mechanism
ClusterProfiler was used to perform the functional enrichment analysis of the KFC targeted genes, we obtained pathways with a P -value < 0.05. Among these pathways, the small lung cancer and NSCLC pathways are highly correlated with the pathogenesis of lung cancer. Increasing evidence confirms that the downstream gene regulation of the PI3K/Akt signaling pathway (hsa04151) can be changed by targeting the GPCR receptor family through PI3K and Akt. Furthermore, activation of the PI3K/Akt signaling pathway can inhibit apoptosis, promote gene transcription, and cell proliferation, accelerate the cell cycle process, and promote angiogenesis by regulating various downstream activating factors [ 61 , 62 , 63 ]. The MAPK signaling pathway (hsa04010) also proved to be vital to the occurrence and development of tumor, and activation of the MAPK signaling pathway may lead to increased proliferation, migration, and invasion of tumor cells [ 64 ].
To further explore the synergistic effects of KFC targets in different pathways, we combined the enrichment pathways as a comprehensive pathway. The comprehensive pathway included small lung cancer (hsa05222), NSCLC (hsa05223), the PI3K/Akt signaling pathway (hsa04151), and the MAPK signaling pathway (hsa04010) (Fig. 9 , Additional Fig. 2 ). In the combined pathways, some genes products sharing multiple pathways are named as cross-talk gene products, and main cascade targeting module merged with CDR-predicted comprehensive pathways. Some cross-talk gene products appear intermediately in merged pathways, including Ras, PKC, Raf1, MEK, ERK, CDK4/6, CyclinD1, AKT, IKK, NF-κB, and NUR77. In this module, Ras and AKT frequently regulates downstream receptors, which were also predicted as cross-talk gene products, such as Raf1, MEK, IKK, NF-κB, and so on. The downstream receptors of cross-talk gene products were reported relevant to proliferation and cell survival in other pathways. Myc and POLK, which have indirect effects on increased survival and cell cycle progression, are still worthy of attention. With the prediction of CDR model and the analysis of merged pathways, KFC could act on these targets and deprive indispensable conditions for the proliferation and long-term survival of cancer cells. In this way, therapeutic mechanism could be achieved possibly.
Main cascade targeting module merged by CDR-predicted cascade pathways. The red units denote the KFC targeted cross-talk genes shared by multiple pathways. The blue units indicate that the KFC targeted genes exist only in one pathway. The white units are annotations or nontargeted proteins
Lung cancer is the most common primary malignancy and ranks first in the incidence and mortality of malignant tumors in the world [ 1 ]. Lung cancer treatments in clinical usually present obvious side effects in clinical use, and can cause serious or general damage to specific or all tissues [ 2 ]. Many clinical studies have shown that anti-lung cancer treatment combined with TCM can reduce the side effects of radiotherapy and chemotherapy, improve therapeutic effects, and reduce complications. Meanwhile, TCM can prolong the life of lung cancer patients and improve their survival quality of life [ 6 ]. For example, SMD can alleviate symptoms of patients and can reduce the inflammatory response [ 10 ], which is widely used in the clinical treatment of lung cancer. Thus, the potential molecular mechanism of SMD in treating lung cancer was studied based on integrated network pharmacology strategy.
In this study, we designed an integrated network pharmacology strategy to explore the KFC of SMD in the treatment of lung cancer and analyze the possible mechanisms. Herbal components were obtained from TCMSP [ 21 ], ETCM [ 22 ], and SymMap [ 23 ], then potential active components were selected by ADMET properties. The targets of these active components were predicted by Hitpick [ 28 ], SwissTargetPrediction [ 27 ], and SEA [ 29 ], then the CT network was constructed. Pathogenic gene data set was downloaded from the DisGeNET. PPI data of pathogenic genes and predicted targets was obtained from STRING [ 30 ] database. Thus, a four-layer component-target-gene-disease network by combining CT, PPI network, and pathogenic gene network, was constructed. With our newly designed node-importance calculation method, key functional network was extracted. Based on knapsack algorithm, KFC were sought out to deduce the maximum target coverage of the key functional network. The accuracy and reliability of the KFC selective method was confirmed further with functional enrichment analysis and in vitro experiments.
Based on integrated network pharmacology strategy, the model designed to explore the KFC of SMD has two measurable advantages. The first is that a novel node-importance calculation model was designed to the figure out key functional networks and validate the coverage rate of key functional networks at functional level. The influence and control of a node to others have been taken into account by the node-importance calculation model, as well as the probability of a node connected to others. The results suggest that the node-importance calculation model we proposed can retain the intervention information to the greatest extent. The second advantage is that we develop the CDR model to capture the KFC in key functional networks. Compared with the traditional pharmacology method based on one-way target speculation mechanisms, the main innovation of our model is that it can provide a methodological reference for the development of integrated pharmacology by considering the spread of the intervention effect from targets to pathogenic genes.
With the prediction of CDR model and the analysis of merged pathways, cross-talk gene products were found, and main cascade targeting module merged with CDR-predicted signalling pathways was visualized clearly. Some cross-talk gene products appear intermediately in merged pathway. Among them, Ras and AKT frequently regulates downstream receptors and other cross-talk gene products, affecting proliferation and cell survival in other pathways. Other cross-talk gene products are still worthy of attention, such as Myc and POLK, despite indirect effects on increased survival and cell cycle progression as reported. After computational analysis and in vitro experiments, KFC could act on these cross-talk gene products to treat lung cancer, and possible therapeutic mechanism could be decoded.
Compared with the traditional pharmacology method based on one-way target speculation mechanisms, the main innovation of our model is that it can provide a methodological reference for the development of integrated pharmacology by considering the spread of the intervention effect from targets to pathogenic genes.
However, it still exists two limitations in our study. First, concentrations of herbal components, that could measure the effects of drug intervention network more exactly, are ignored in our network pharmacological analysis. Second, in order to validate the reliability of our approach, more KFC should be selected to determine the effect of viability of A549 cells or in vivo experiments.
In conclusion, the molecular mechanism of SMD treatment of lung cancer was revealed by integrated network pharmacology model and experimental validation. The strategy proposed in this study can be used to identify key compounds in the complex network and provides an operable test range for subsequent experimental verification, which greatly reduces the experimental workload. In follow-up studies, in vitro and in vivo animal studies can be designed based on the predicted KFC.
Availability of data and materials
All data included in this study are available upon request by contact with the corresponding author. Human data in this study was obtained from public database DisGeNET ( https://www.disgenet.org/ ).
Abbreviations
Absorption, Distribution, Metabolism, Excretion, and Toxicity
Contribution Decision Rate
Contribution Index
Components-Targets-Pathogenetic genes
Drug-Likeness
Dulbecco’s Modified Eagle’s Medium
Fetal Bovine Serum
Gene Ontology
Glehnia littoralis (A.Gray) F.Schmidt ex Miq.
Glycyrrhiza uralensis Fisch. ex DC.
International Classification of Diseases
Key Functional Components
Kyoto Encyclopedia of Genes and Genomes
Lablab purpureus Subsp. purpureus
Morus alba L.
Non-Small Cell Lung Cancer
Ophiopogon japonicus (Thunb.) Ker Gawl.
Oral Bioavailability
Polygonatum odoratum (Mill.) Druce
Protein–Protein Interactions
Shashen Maidong Decoction
Similarity Ensemble Approach
Symptom Mapping
Traditional Chinese Medicine
Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform
Trichosanthes kirilowii Maxim
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Article PubMed Google Scholar
Ortiz-Cuaran S, Scheffler M, Plenker D, et al. Heterogeneous mechanisms of primary and acquired resistance to third-generation EGFR inhibitors. Clin Cancer Res. 2016;22(19):4837–47.
Article CAS PubMed Google Scholar
Lin YT, Chen JS, Liao WY, et al. Clinical outcomes and secondary epidermal growth factor receptor (EGFR) T790M mutation among first-line gefitinib, erlotinib and afatinib-treated non-small cell lung cancer patients with activating EGFR mutations. Int J Cancer. 2019;144(11):2887–96.
Cheng G, Zhang L. Adverse events related to bevacizumab and the management principles in non-small cell lung cancer. Zhongguo Fei Ai Za Zhi. 2010;13(6):563–7.
CAS PubMed Google Scholar
Zhao Z, Ni Y, Li L, Xin T. Acquired drug resistance mechanism of osimertinib in the targeted therapy of non-small cell lung cancer. Zhongguo Fei Ai Za Zhi. 2020;23(4):274–81.
PubMed Google Scholar
Su XL, Wang JW, Che H, et al. Clinical application and mechanism of traditional Chinese medicine in treatment of lung cancer. Chin Med J (Engl). 2020;133(24):2987–97.
Zhao B, Hui X, Jiao L, et al. A TCM formula YYWY inhibits tumor growth in non-small cell lung cancer and enhances immune-response through facilitating the maturation of dendritic cells. Front Pharmacol. 2020;11:798.
Article CAS PubMed PubMed Central Google Scholar
Zheng X, Wang W, Wang G, Liu S. Could Jinfukang alleviate the chemotherapy-related adverse effects in non-small cell lung cancer patients?: A protocol for a double-blind, randomized controlled trial. Medicine (Baltimore). 2021;100(28):e25002.
Xu ZY, Jin CJ, Zhou CC, et al. Treatment of advanced non-small-cell lung cancer with Chinese herbal medicine by stages combined with chemotherapy. J Cancer Res Clin Oncol. 2011;137(7):1117–22.
He M, Luo Y, Chen L, et al. Shashen maidong decoction: the effect of TNF-alpha and IL-6 on lung cancer cachexia based on cancer toxicity theory. Am J Transl Res. 2021;13(6):6752–8.
CAS PubMed PubMed Central Google Scholar
Huang QH, Liang YJ, Chen N, Du XT. Effect of Shashen Maidong decoction on tumor marker levels in patients with non-small cell lung cancer. Mod Med Health Res Electron J. 2021;5(12):87–9.
Google Scholar
Wang Y, Yang SY, Pan YN, et al. lnfluences of Shashen Maidong decoction-containing serum combined with cisplatin on expressions of E-cadherin and snail in lung adenocarcinoma A549 cells. Chin Arch Tradit Chin Med. 2021;39(08):62–5.
Xu WW, Liu CY. Effect of Shashen Maidong decoction combined with cisplatin on proliferation of lung cancer cell line A549 and expressions of Smad7 and TGF-β1 protein. Liaoning J Tradit Chin Med. 2020;47(05):144–7.
CAS Google Scholar
Chen J, Yuan J, Zhou L, et al. Regulation of different components from ophiopogon japonicus on autophagy in human lung adenocarcinoma A549Cells through PI3K/Akt/mTOR signaling pathway. Biomed Pharmacother. 2017;87:118–26.
Wu J, Gao W, Song Z, et al. Anticancer activity of polysaccharide from Glehnia littoralis on human lung cancer cell line A549. Int J Biol Macromol. 2018;106:464–72.
Li S, Zhou X, Xiong H. Clinical study of Shashenmaidong decoction in the treatment of clinical study of Shashenmaidong decoction in the treatment of clinical study of Shashenmaidong decoction in the treatment of clinical study of Shashenmaidong decoction in the treatment of radiation pneumonitis. Acta Chin Med. 2015;30(03):328–9.
Liu M, Wu YL, Zeng R. Observation on the effect of Jiajian Shashen Maidong decoction combined with chemotherapy in the treatment of non-small cell lung cancer. Chin J Clin Rational Drug Use. 2020;13(11):53–4.
Wang KX, Gao Y, Lu C, et al. Uncovering the complexity mechanism of different formulas treatment for rheumatoid arthritis based on a novel network pharmacology model. Front Pharmacol. 2020;11:1035.
Yang L, Fan L, Wang K, et al. Analysis of molecular mechanism of erxian decoction in treating osteoporosis based on formula optimization model. Oxid Med Cell Longev. 2021;2021:6641838.
PubMed PubMed Central Google Scholar
Pinero J, Ramirez-Anguita JM, Sauch-Pitarch J, et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48(D1):D845–55.
Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13.
Article PubMed PubMed Central Google Scholar
Xu HY, Zhang YQ, Liu ZM, et al. ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic Acids Res. 2019;47(D1):D976–82.
Wu Y, Zhang F, Yang K, et al. SymMap: an integrative database of traditional Chinese medicine enhanced by symptom mapping. Nucleic Acids Res. 2019;47(D1):D1110–7.
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J Cheminform. 2011;3:33.
Benet LZ, Hosey CM, Ursu O, Oprea TI. BDDCS, the rule of 5 and drugability. Adv Drug Deliv Rev. 2016;101:89–98.
Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717.
Gfeller D, Grosdidier A, Wirth M, Daina A, Michielin O, Zoete V. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014;42(Web Server issue):W32–8.
Liu X, Vogt I, Haque T, Campillos M. HitPick: a web server for hit identification and target prediction of chemical screenings. Bioinformatics. 2013;29(15):1910–2.
Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25(2):197–206.
Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–13.
Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–7.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28(11):1947–51.
Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51(D1):D587–92.
Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag; 2016.
Lv J, Zhu S, Chen H, et al. Paeonol inhibits human lung cancer cell viability and metastasis in vitro via miR-126-5p/ZEB2 axis. Drug Dev Res. 2022;83(2):432–46. https://doi.org/10.1002/ddr.21873 .
Zhang L, Chen WX, Li LL, et al. Paeonol suppresses proliferation and motility of non-small-cell lung cancer cells by disrupting STAT3/NF-kappaB signaling. Front Pharmacol. 2020;11:572616.
Lin CL, Chen RF, Chen JY, et al. Protective effect of caffeic acid on paclitaxel induced anti-proliferation and apoptosis of lung cancer cells involves NF-kappaB pathway. Int J Mol Sci. 2012;13(5):6236–45.
Yin MC, Lin CC, Wu HC, Tsao SM, Hsu CK. Apoptotic effects of protocatechuic acid in human breast, lung, liver, cervix, and prostate cancer cells: potential mechanisms of action. J Agric Food Chem. 2009;57(14):6468–73.
Sun Y. Chemical composition analysis and pharmacokinetic study of the Shasheng Maidong decoction. PhD thesis. Hebei Medical University; 2016.
Khan F, Niaz K, Maqbool F, et al. Molecular targets underlying the anticancer effects of quercetin: an update. Nutrients. 2016;8(9):529.
Wu F, Chen J, Fan LM, et al. Analysis of the effect of rutin on GSK-3beta and TNF-alpha expression in lung cancer. Exp Ther Med. 2017;14(1):127–30.
Tian Q, Wang L, Sun X, Zeng F, Pan Q, Xue M. Scopoletin exerts anticancer effects on human cervical cancer cell lines by triggering apoptosis, cell cycle arrest, inhibition of cell invasion and PI3K/AKT signalling pathway. J BUON. 2019;24(3):997–1002.
Velli SK, Sundaram J, Murugan M, Balaraman G, Thiruvengadam D. Protective effect of vanillic acid against benzo(a)pyrene induced lung cancer in Swiss albino mice. J Biochem Mol Toxicol. 2019;33(10):e22382.
Reczynska K, Marchwica P, Khanal D, et al. Stimuli-sensitive fatty acid-based microparticles for the treatment of lung cancer. Mater Sci Eng C Mater Biol Appl. 2020;111:110801.
Cai Y, Jia R, Xiong H, et al. Integrative gene expression profiling reveals that dysregulated triple microRNAs confer paclitaxel resistance in non-small cell lung cancer via co-targeting MAPT. Cancer Manag Res. 2019;11:7391–404.
Mokhlesi A, Talkhabi M. Comprehensive transcriptomic analysis identifies novel regulators of lung adenocarcinoma. J Cell Commun Signal. 2020;14(4):453–65.
Khomenko TM, Zakharenko AL, Chepanova AA, et al. Promising new inhibitors of Tyrosyl-DNA phosphodiesterase I (Tdp 1) combining 4-arylcoumarin and monoterpenoid moieties as components of complex antitumor therapy. Int J Mol Sci. 2019;21(1):126.
Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361(10):958–67.
Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–39.
Agca CA, Kirici M, Nedzvetsky VS, Gundogdu R, Tykhomyrov AA. The effect of TIGAR knockdown on apoptotic and epithelial-mesenchymal markers expression in doxorubicin-resistant non-small cell lung cancer A549 cell lines. Chem Biodivers. 2020;17(9):e2000441.
Manabe S, Kasajima R, Murakami S, et al. Analysis of targeted somatic mutations in pleomorphic carcinoma of the lung using next-generation sequencing technique. Thoracic Cancer. 2020;11(8):2262–9.
Yang W, You N, Jia M, et al. Undetectable circulating tumor DNA levels correlate with low risk of recurrence/metastasis in postoperative pathologic stage I lung adenocarcinoma patients. Lung Cancer. 2020;146:327–34.
Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. Lancet. 2013;382(9893):720–31.
Chen JA, Riess JW. Optimal management of patients with advanced NSCLC harboring high PD-L1 expression and driver mutations. Curr Treat Options Oncol. 2020;21(7):60.
Peters S, Camidge DR, Shaw AT, et al. Alectinib versus crizotinib in untreated ALK-positive non-small-cell lung cancer. N Engl J Med. 2017;377(9):829–38.
Verma P, Ghosh A, Ray M, Sarkar S. Lauric acid modulates cancer-associated microRNA expression and inhibits the growth of the cancer cell. Anticancer Agents Med Chem. 2020;20(7):834–44.
Kirkland JB. Niacin requirements for genomic stability. Mutat Res. 2012;733(1–2):14–20.
Setia MS. Methodology series module 5: sampling strategies. Indian J Dermatol. 2016;61(5):505–9.
Wang C, Li S, Liu J, et al. Silencing of S-phase kinase-associated protein 2 enhances radiosensitivity of esophageal cancer cells through inhibition of PI3K/AKT signaling pathway. Genomics. 2020;112(5):3504–10.
Ueda K, Nakahara T, Akanuma K, Mori A, Sakamoto K, Ishii K. Differential effects of LY294002 and wortmannin on neurons and vascular endothelial cells in the rat retina. Pharmacol Rep. 2013;65(4):854–62.
Yang P, Zhao J, Hou L, Yang L, Wu K, Zhang L. Vitamin E succinate induces apoptosis via the PI3K/AKT signaling pathways in EC109 esophageal cancer cells. Mol Med Rep. 2016;14(2):1531–7.
Li C, Liu DR, Li GG, et al. CD97 promotes gastric cancer cell proliferation and invasion through exosome-mediated MAPK signaling pathway. World J Gastroenterol. 2015;21(20):6215–28.
Download references
Acknowledgements
Not applicable.
This study was financially supported by the Startup fund from the Southern Medical University [grant No. G820282016], the Natural Science Foundation Council of China [grant No. 31501080, 32070676], Natural Science Foundation of Guangdong Province [grant No. 2021A1515010737], Hong Kong Baptist University Strategic Development Fund [grant No. SDF13-1209-P01, SDF15-0324-P02(b) and SDF19-0402-P02], Hong Kong Baptist University Interdisciplinary Research Matching Scheme [grant No. RC/IRCs/17–18/04].
Author information
Jieqi Cai, Yupeng Chen and Kexin Wang contributed equally to this work.
Authors and Affiliations
Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Southern Medical University, Guangzhou, Guangdong Province, China
Jieqi Cai, Yupeng Chen, Yi Li, Jie Wu, Hailang Yu, Wei Meng, Handuo Wang & Daogang Guan
Guangdong Provincial Key Laboratory of Single Cell Technology and Application, Guangzhou, Guangdong Province, China
Neurosurgery Center, Guangdong Provincial Key Laboratory on Brain Function Repair and Regeneration, Department of Cerebrovascular Surgery, Engineering Technology Research Center of Education Ministry of China on Diagnosis and Treatment of Cerebrovascular Disease, Zhujiang Hospital, Southern Medical University, Guangzhou, Guangdong, 510280, China
Division of Hepatobiliopancreatic Surgery, Department of General Surgery, Nanfang Hospital, Southern Medical University, Guangzhou, Guangdong, China
Qingping Li
Department of Burns, Nanfang Hospital, Southern Medical University, Guangzhou, China
Institute of Integrated Bioinformedicine and Translational Science, Hong Kong Baptist University, Hong Kong, China
Department of Histology and Embryology, School of Basic Medical Sciences, Southern Medical University, Guangzhou, Guangdong Province, China
Mianbo Huang
Huiqiao Medical Center, Nanfang Hospital, Southern Medical University, Guangzhou, China
You can also search for this author in PubMed Google Scholar
Contributions
GXW, MBH, and DGG provided the research concept and designed the study. JQC, YPC, and KXW conducted the analyses and wrote the manuscript. YL, JW, and WM participated in data analysis. HLY, QPL, QW and HDW conducted experiments. APL, MBH and DGG contributed to revising and proof-reading the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Correspondence to Mianbo Huang , Genxia Wei or Daogang Guan .
Ethics declarations
Ethics approval and consent to participate.
Human lung cancer A549 cells were obtained from GuangZhou Jennio Biotech Co., Ltd. The ethics committee of Southern Medical University confirms that this study would have had the need for ethics approval waived.
Consent for publication
Competing interests.
The authors declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1..
Supporting Information.
Additional file 2: Additional Table 1.
All herbal components of SMD. Additional Table 2. Active components of SMD. Additional Table 3. Predicted targets of active components from SMD. Additional Table 4. Pathogenic genes of lung cancer downloaded from DisGeNET database. Additional Table 5. The CDR of active components.
Additional file 3: Additional Figure 1.
Construct complex components-targets-pathogenic genes-disease (C-T-P-D) network. Green units represent active components of SMD, blue units represent predicted targets of active components and red units represent predicted pathogenic genes of lung cancer. The gray lines indicate the interactions. Additional Figure 2. Full distribution of KFC targets on merged paths. The red units denote the KFC targeted cross-talk genes shared by multiple pathways, and the red dashed line connects them together in different pathways. The blue units indicate that the KFC targeted genes exist only in one pathway. The white units are annotations or nontargeted proteins.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Cai, J., Chen, Y., Wang, K. et al. Decoding the key compounds and mechanism of Shashen Maidong decoction in the treatment of lung cancer. BMC Complement Med Ther 23 , 158 (2023). https://doi.org/10.1186/s12906-023-03985-y
Download citation
Received : 14 September 2022
Accepted : 29 April 2023
Published : 15 May 2023
DOI : https://doi.org/10.1186/s12906-023-03985-y
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Shashen Maidong decoction
- Lung cancer
- Network analysis
- Key functional networks
- Contribution decision rate
BMC Complementary Medicine and Therapies
ISSN: 2662-7671
- Submission enquiries: [email protected]
- General enquiries: [email protected]
Thesis Defense – Jessica Petrochuk
May 1 @ 12:00 pm - 1:00 pm.

Thesis Defense: Jessica Petrochuk
Time: Monday May 1, 2024, at 12:00 PM Eastern Time
Location: Kresge 205 Zoom: https://harvard.zoom.us/j/97662851929
Jessica will present the thesis titled “Foundation Model Based Prediction of Lung Cancer Survival Using Temporal Changes in Dual Time Point CT Scans”. The thesis committee is chaired by Raymond Mak and includes Benjamin Kann and John Quackenbush.
- Google Calendar
- Outlook 365
- Outlook Live
Related Events

Thesis Defense – Evelyn Jiang

Thesis Defense – Yushan Xu

Thesis Defense – Jiaxin Shen
News from the school.

From public servant to public health student

Exploring the intersection of health, mindfulness, and climate change

Conference aims to help experts foster health equity

Building solidarity to face global injustice
- Current Article
First annual PICR Awards ceremony recognizes excellence in cancer research

In a significant milestone for the Purdue Institute for Cancer Research (PICR), faculty, students and staff gathered at the Hansen Life Sciences Research Building on April 17 to honor seven awardees for their outstanding contributions to cancer research. PICR Director Andrew Mesecar delivered remarks emphasizing the importance of establishing the awards ceremony as a tradition and spoke about the distinct contributions each recipient had made to further the institute’s goal of conquering cancer.
“In almost every nomination and support letter we received, a single word stood out — and that word was ‘excellence.’ From scientific achievements to service within our cancer research community, the word excellence was used to describe the contributions of each of our nominees and awardees. Excellence describes what we strive to achieve in all areas of PICR’s mission, from research to training and education to inclusion and belonging,” Mesecar said. He also emphasized that “through the dedicated efforts of over 115 faculty members and more than 600 students and post-docs, the Purdue Institute for Cancer Research is indeed delivering on President Chiang’s vision of excellence at scale.”
Get to know the PICR Award recipients
Trainee awards.
These awards are presented to trainees who exhibit outstanding commitment, creativity and excellence in cancer research, and demonstrate exceptional promise for productive careers that will have meaningful impact in their fields.
Outstanding Undergraduate Student Award
A senior in the College of Pharmacy majoring in pharmaceutical sciences, Sam King is a member of the Honors College and president of the Undergraduate Research Society. King has been an undergraduate research assistant in Emily Dykhuizen’s lab since 2021. He will graduate with a bachelor’s degree this May.
Outstanding Doctoral Student Award
Luopin wang.
A PhD candidate in the biochemistry and computer science departments, Luopin Wang has been a graduate research assistant in Majid Kazemian’s lab since 2018. Now in the final year of her doctoral program, Wang passed her dissertation defense and will be graduating in May with a PhD in computer science.
Outstanding Postdoctoral Fellow Award
Ikjot singh sohal.
Ikjot Singh Sohal received his PhD in biomedical engineering and biotechnology from the University of Massachusetts and has been a postdoctoral research associate in Andrea Kasinski’s lab since 2018. His research focuses on understanding the biogenesis of extracellular vesicles and their role in immune suppression, as well as developing targeted miRNA therapeutics in lung and prostate cancer.
Outstanding Service and Engagement Award
Doug cuttell, managing director, picr.
This award recognizes and celebrates individuals who have demonstrated exceptional commitment and active participation within the institute. It honors PICR members, staff, trainees or volunteers who have not only contributed significantly to the goals and mission of the institute but have also actively engaged in its various activities, fostering a collaborative and impactful environment.
Faculty Awards
Outstanding early career research achievement award, xiaoping bao.
This award recognizes early career research that goes beyond the awardee’s dissertation and demonstrates clear scientific or scholarly contributions in work completed as a PICR member. The award also acknowledges meaningful impact on their field.
Xiaoping Bao is the William K. Luckow Assistant Professor in the Davidson School of Chemical Engineering. His research focuses on stem cell immuno-engineering with cancer as the target disease. He has worked on the development of chimeric antigen receptor neutrophils for use in targeted cancer therapy.
Outstanding Mid-Career Research Achievement Award
Andrea kasinski, deputy director, picr.
This award recognizes strong scientific contributions by a mid-career PICR member. The award acknowledges sustained and significant impact on their field and national or international recognition for their work. As PICR deputy director and associate professor of biological sciences, Andrea Kasinski’s research focuses on identifying roles for microRNAs in cancer development and progression, and applying this knowledge in clinical applications.
Outstanding Career Research Achievement Award
John tesmer.
This award recognizes tenured full professors who have made strong scientific or scholarly contributions in their work as a PICR member. The award honors sustained and significant impact on their field, and national and international recognition for their work.
John Tesmer, the Walther Distinguished Professor in Cancer Structural Biology in the Department of Biological Sciences and professor of medicinal chemistry and molecular pharmacology, is a leader of PICR’s Targets, Structures and Drugs scientific program. His research focuses on structure, function and chemical-biological targeting of eukaryotic signaling proteins and cholesterol metabolizing enzymes.
Next year, PICR will introduce awards for graduate students and/or post-docs and faculty to recognize mentoring and training.
Learn more about the PICR award opportunities here .
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Front Cell Infect Microbiol
- PMC10167031
The traditional Chinese medicine and non-small cell lung cancer: from a gut microbiome perspective
Xuelin wang.
1 School of Food Science and Engineering (School of Biological and Pharmaceutical Sciences), Shaanxi University of Science & Technology, Xi an, China
2 Department of Geriatrics, Xijing Hospital, Fourth Military Medical University, Xi an, China
Mengzhou Wang
Non-small cell lung cancer (NSCLC) is one of the most serious diseases affecting human health today, and current research is focusing on gut flora. There is a correlation between intestinal flora imbalance and lung cancer, but the specific mechanism is not clear. Based on the “lung and large intestine being interior-exteriorly related” and the “lung-intestinal axis” theory. Here, based on the theoretical comparisons of Chinese and western medicine, we summarized the regulation of intestinal flora in NSCLC by active ingredients of traditional Chinese medicine and Chinese herbal compounds and their intervention effects, which is conducive to providing new strategies and ideas for clinical prevention and treatment of NSCLC.
1. Introduction
In the womb, the fetus begins to develop its gut microbiota, and an estimated 40 trillion microorganisms are considered to reside on and in the human body. The digestive tract, often known as the “gut microbiome (GM),” contains the most microbial species variety. ( Nagasaka et al., 2020 ). Although its primary function has been thought to be to protect against pathogen overgrowth in the gut, the gut microbiome appears to play a critical role in the maturation and ongoing education of the host immune response and likely has significant effects in many conditions not typically considered infectious diseases ( Fulde and Hornef, 2014 ; Kamada et al., 2013 ). Intestinal microbiome imbalances, also known as ecological imbalances, have been linked to a variety of illnesses, including cancer, in recent years. Given that the gut microbiome is constantly exposed to a wide spectrum of potential pathogens, it is not surprising that it is vital to the host immune response.
According to the latest data from China’s National Cancer Center in 2022, lung cancer is the second most common cancer in humans worldwide, with the highest incidence of morbidity and mortality. NSCLC accounts for more than 80% of all lung cancers, and it is one of the most challenging to treat with a poor response to immune checkpoints (ICIs) in most patients. Interestingly, emerging evidence has suggested that microbiota may also play vital roles in lung cancers at multiple levels ( Vernocchi et al., 2020 ).
Additionally, there is mounting evidence linking the GM and its metabolome to the response to ICI treatment in NSCLC ( Jin et al., 2019 ; Botticelli et al., 2020 ; Hakozaki et al., 2020 ). In fact, the metabolites of the microbes as well as their cells contribute to the stimulation of the immune response. Their interactions stimulate and trigger an immunological response, helping the host immune system combat cancer. The relationship between the gut microbiota and non-small cell lung cancer will therefore be the main topic of this essay.
2. Close relationship: GM and NSCLC
2.1. the overview of gm.
A wide variety of microorganisms, including bacteria, fungus, archae, and viruses, live in the complex, dynamic, and geographically heterogeneous ecosystem known as the human GM ( Chen et al., 2021b ). The total genetic repertory of all gut microbes is an order of magnitude greater than the genetic repertoire of the human genome, and also encodes many more unique genes than the host genome, and they generate more than 1000 metabolites ( Fan and Pedersen, 2021 ). It is also referred to as the “essential organ” of the human body ( Ding et al., 2019 ). The GM is the body’s greatest micro-ecosystem and works in symbiosis with the host to sustain regular physiological functions in a state of dynamic balance ( Figure 1 ). Human host receives a number of crucial services from the gut microbiota attests to its significance, including the conversion of indigestible dietary components, the creation of vital vitamins, the elimination of harmful substances, the defeat of infections, the augmentation of the intestinal barrier, and the stimulation and control of the immune system ( Heintz-Buschart and Wilmes, 2018 ), these are necessary to support normal tissue and organ function. It is generally known that the gut microbiota directly affects both health and disease status. The microbiome has a significant impact on host physiology due to its broad metabolic and synthetic capabilities and its intricate interactions with the formation and control of the host immune system ( Walker et al., 2021 ). Despite the symbiotic nature of the intestinal host-microbial relationship, the close association of an abundant bacterial community with intestinal tissues poses immense health challenges ( Hooper et al., 2012 ).

The gut microbiome contains the greatest diversity of microbial species, and plays a critical role in the immunoregulation, antibacterial polypeptide, anti-adhesive,the intestinal barrier,and so on.
2.2. The composition of GM
The development of the technique to sequence the bacterial 16S ribosomal RNA gene allowed overall taxonomic assessment of the gut microbiome, and this has dramatically increased our knowledge of the broad variations in microbial composition ( Weersma et al., 2020 ). The human gut microbiota consists of several types of microbes, including bacteria, archaea, eukarya, viruses, and parasites. More than 1000 species of bacteria have been identified in the human gut, although a person on average only carries 160 species ( Berding et al., 2021 ). The gut microenvironment mainly favors the growth of bacteria from seven predominant divisions (Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteria, Proteobacteria, Verrucomicrobia, and Cyanobacteria). Among these seven divisions, the Bacteroidetes and Firmicutes constitute more than 90% of the total population ( Adak and Khan, 2019 ). Studies indicate links between dysbiosis or disturbance in the microbiome and diseases that not only affect the gut but also organs like the lung, thyroid, brain, cardiac, immune system, etc. ( Tang et al., 2019 ; Hufnagl et al., 2020 ; Knezevic et al., 2020 ; Rutsch et al., 2020 ; Wang et al., 2022a ). The crosstalk between the gut microbiome and distal organs is being increasingly recognized, and host-microbiome interactions are being delineated piece by piece. Gut microbes and their associated metabolites are thought to cause and modulate lung cancer development, albeit influenced by the host genetic make-up and environment. Non-targeted metabolomics approach based on LC-MS can successfully distinguish lung cancer patients from healthy individuals. Also, the microbial diversity in lung cancer patients is significantly higher than that of normal individuals ( Zhao et al., 2021 ).
2.3. The role of GM in NSCLC (Lung-gut axis)
Chinese medicine believes that “the lung and large intestine are interior-exteriorly related” ( Ni et al., 2022 ). The hypothesis of the lung-gut axis, which modern medicine has advanced, corresponds to the Chinese medical theory of the “lung and large intestine being interior-exteriorly related.” The lungs and large intestine can work together to modulate immunity and inflammation through the lung-gut axis, in which the movement of the gut microbiota and metabolites is the most important communication mechanism ( Figure 2 ). The lungs and the gut both develop from the same embryo. The gut and the lungs, like all other organs included in the MIS compartments, are mucosally similar, promoting comparable dynamics in the interactions between the immune system and their microbiota. Moreover, they are indirectly connected via the circulatory and lymphatic systems ( Pizzo et al., 2022 ). The lungs do indeed have a specific microbiota. The predominant bacterial phyla in the lungs of healthy subjects are the same as those in the gut. These are mainly Firmicutes (Staphylococcus, La-ctobacillus, and Streptococcus) and Bacteroidetes, followed by Proteobacteria and Actinobacteria ( Georgiou et al., 2021 ). While lung cancer patients present higher levels of Bacteroidetes, Fusobacteria, Cyanobacteria, Spirochaetes, and Lentisphaerae, and lower levels of Bacteroidetes, Firmicutes, and Verrucomicrobia in their lung and gut microbiota ( Corrêa et al., 2022 ).

The lungs and large intestine can work together to modulate immunity and inflammation through the lung-gut axis, in which the movement of the gut microbiota and metabolites is the most important communication mechanism. Moreover, they are indirectly connected via the circulatory and lymphatic systems.
Although the gastrointestinal and respiratory systems are separated by physical distance, they share a common embryonic origin and exhibit a striking structural similarity, suggesting the possibility of multimodal interaction between these two tracts. As a result, the gut-lung axis, a new and distinct interaction between the respiratory and gastrointestinal tracts, has been created. According to reports, this two-way regulation of the gut-lung axis organs is accomplished through microbial and immunological processes. A growing body of research suggests that the microbiome is crucial in inflammatory pulmonary disorders such as acute lung injury (ALI) and acute respiratory distress syndrome. Tang et al. discovered that the transition from ulcerative colitis (UC) to colorectal cancer (CRC) significantly altered not only the composition of the gut microbiota and metabolites associated with inflammation but also the lung tissues, which demonstrated that gastrointestinal illnesses can result in pulmonary illnesses ( Tang et al., 2022 ). Yoon et al. found that the composition of the gut microbiota has a significant impact on BLM-induced wasting and death, suggesting a role for the lung-gut axis in lung injury. They also found that the presence of specific gut commensal microbes may be a risk factor for having more severe inflammatory lung diseases ( Yoon et al., 2022 ).
2.3.1. GM regulated inflammation and immune system
It’s vital to keep in mind that inflammation can have two opposing impacts on tumors: whereas local inflammation limited to the tumor microenvironment can reduce the tumor, chronic, broad inflammation generally promotes tumor growth. According to preclinical research in mouse models ( Sánchez-Alcoholado et al., 2020 ), GM-mediated colorectal cancer (CRC) and inflammation have a high correlation. Guo et al. revealed that Ganoderma lucidum (GLP) decreased colitis and tumorigenesis. The potential explanation is that GLP ameliorated microbiota dysbiosis, increased short-chain fatty acid production, profoundly improved gut barrier function as evidenced by increased numbers of goblet cells, MUC2 secretion, and tight junction protein expressions. Simultaneously, GLP treatment inhibited macrophage infiltration and downregulated IL-1β, iNOS, and COX-2 expressions ( Guo et al., 2021a ).
The gut microbiome plays a key role in intestinal permeability and immune regulation. The gut microbiome regulates immune cell populations in part through short-chain fatty acids, which can restore colonic regulatory T cell populations in germ-free mice and signalling via Toll-like receptors (TLRs) among other innate and adaptive immune pathways ( Leigh and Morris, 2020 ). Peng et al. recruited 74 patients with advanced-stage gastrointestinal (GI) cancer receiving anti-PD-1/PD-L1 treatment and collected their fecal samples prior to and during immunotherapy, along with clinical evaluations. They revealed an elevation of the Prevotella/Bacteroides ratio in patients, with a preferred response to antiPD-1/PD-L1 treatment and a particular subgroup of responders harboring a significantly higher abundance of Prevotella, Ruminococcaceae , and Lachnospiraceae ( Peng et al., 2020 ). Huang et al. found Ginseng polysaccharides (GPs) increased the antitumour response to αPD-1 monoclonal antibody (mAb) by increasing the microbial metabolites valeric acid and decreasing L-kynurenine, as well as the ratio of kynurenine/tryptophan, which contributed to the suppression of regulatory T cells and induction of Teff cells after combination treatment. And the microbial analysis indicated that the abundance of Parabacteroides distasonis and Bacteroides vulgatus was higher in responders to anti-PD-1 blockade than in non-responders in the clinic( Huang et al., 2022 ). And a clinical study of 37 patients with advanced NSCLC in China reveal strong correlation between gut microbiome diversity and the responses to anti-PD-1 immunotherapy. Patients with a favorable gut microbiome exhibit enhanced memory T cell and natural killer cell signatures in the periphery ( Jin et al., 2019 ).
2.3.2. Metabolites of GM in NSCLC
The short-chain fatty acids (SCFAs), which are the major metabolic products of the GM from dietary fiber (especially in the case of a high-fiber diet), are key mediators of the host–microbiome interaction and perform countless functions with localized and systemic effects. Main SCFAs with total intestinal concentrations exceeding 100 mM include propionate, acetate, and butyrate ( Mirzaei et al., 2021 ). The basic function of these fatty acids is to provide energy. SCFAs also act as signaling molecules by mediating metabolic processes and immune responses, and various studies have proven their impressive anti-inflammatory action and antitumor potential. One study showed that sodium butyrate affects proliferation and migration of A549 cells by activating the TNF receptor-associated factor 6 (TRAF6)-thioredoxin-interacting protein (TXNIP) pathway, suggesting that sodium butyrate has an effective therapeutic effect on lung adenocarcinoma. ( Xiao et al., 2020 ). It was also found that sodium butyrate and docetaxel alone, respectively, inhibited proliferation and promoted apoptosis in A549 cells in vitro and in vivo . Furthermore, the combined therapy decreased protein expression of Ki-67, CDK1, CDK2, Cyclin D1, Bcl-2, and Survivin and increased protein expression of Cyclin A, p21, Bax and cleaved-Caspase 3 ( Chen et al., 2020b ). Chen et al. reported that propionate and butyrate produced by gut microbiota after probiotic supplementation can attenuate the lung metastasis of melanoma cells in mice ( Chen et al., 2021a ). Sodium propionate (SP) inhibited lung cancer cell proliferation by inducing cell apoptosis and cell cycle arrest, especially in the G2/M phase ( Kim et al., 2019 ).
2.3.3. GM involved in the treatment of NSCLC
Unusual bacterial clusters were discovered in the NSCLC patients in an observational investigation with exploratory GM analysis, and the individuals who did not have cachexia were enriched with healthy bacteria at the genus level, including Eubacterium, Anaerostipes, and Blautia ( Hakozaki et al., 2022 ). Akkermansia bacteria, specifically Akkermansia mucinifla, is now being found in relation to supporting therapy and markers for an immunotherapy response in cancer patients. It has been shown that this bacterium improves response to treatment in NSCLC patients receiving immune checkpoint inhibitors (ICIs). In the 47 NSCLC patients studied by Grenda et al., patients with disease stabilization and partial immunotherapy responses had a higher percentage of Akkermansiaceae than patients with cancer progression. Additionally, they discovered that patients with squamous cell carcinoma had more Akkermansiaceae than those with adenocarcinoma. As a result, they suggested that Akkermansiaceae may serve as a supportive marker for NSCLC patients’ immunotherapy responses ( Grenda et al., 2022 ). An animal research indicated that Akkermansia muciniphila (Akk) combining with cisplatin (CDDP) slowed down the growth of tumor volume and improved the changes in tumor pathomorphology and was related to those pathways, including the cytokine-cytokine receptor interaction, Th17 cell differentiation, FOXO, JAK-STAT, and PI3K-Akt signaling pathways ( Chen et al., 2020c ).
Cancer immunotherapy has become highly successful against an array of distinct hematological and solid metastatic malignancies. Immune checkpoint inhibitors (ICIs) targeting the programmed death-1/programmed death-ligand 1 (PD-1/PD-L1) axis induce sustained clinical responses in a measure of cancer patients. Routy et al. found that primary resistance to ICIs can be attributed to abnormal gut microbiome composition ( Routy et al., 2018 ). Their results prove that Fecal microbiota transplantation (FMT) from cancer patients who responded to ICIs into germ-free or antibiotic-treated mice ameliorated the antitumor effects of PD-1 blockade, whereas FMT from nonresponding patients failed to do so. Drug regimens for many tumors also relate to intestinal microecology, such as one of the most popular chemotherapy drugs, paclitaxel (PTX), which was used to treat a variety of tumor types but whoes debilitating side effects included gastrointestinal and behavioral disorders, restricting its use while a 30-day sodium butyrate (BuNa) pre-treatment repaired the altered gut barrier integrity and microbiota composition caused by the PTX. These findings suggest that dietary supplementation with this secure postbiotic may be taken into account for treating PTX-induced central side effects when treating cancer ( Cristiano et al., 2022 ). In addition, probiotic use was linked to better clinical outcomes in patients with advanced or recurrent NSCLC who received anti-PD-1 monotherapy, according to a multicenter retrospective analysis ( Takada et al., 2021 ), which indicates that probiotics may be an superior option for NSCLC patients who receive ICIs.
3. Traditional Chinese medicines and GM
3.1. active components of tcms, 3.1.1. polysaccharide.
Drug therapy using natural substances is currently considered a promising future alternative to traditional medicine. As an important class of biologically active natural products, polysaccharides from TCM play an important role in the field of medicine, including gut microbiome regulation ( Table 1 ), immune regulation, as well as anti-tumor, anti-oxidation, etc. ( Yu et al., 2018 ). Luo et al., who demonstrated that Polygonatum sibiricum polysaccharides-1 (PSP-1) reconstructed the gut microbiota composition, including reducing the relative abundance of Helicobacter, and increasing Akkermansia muciniphila, and revealed that PSP-1 may improve the inflammatory environment and reduce Amyloid-β (Aβ) deposition in the intestine of 5xFAD mice by acting on the bacteria ( Luo et al., 2022 ). Sun et al. found that a water-insoluble polysaccharide (WIP) isolated and identified from the Poria cocos mushroom significantly enhanced the butyrate-producing bacteria Lachnospiracea and Clostridium. It was also demonstrated that WIP treatment increased butyrate-producing in the gut, maintained intestinal integrity, and reducted of endoxemia, and activated the intestinal PPAR-γ pathway ( Sun et al., 2019 ). Some scholars have carried out related research on Astragalus polysaccharide (APS). Zhong et al. indicated that Astragalus mongholicus polysaccharides (mAPS) significantly reduced the Firmicutes to Bacteroidetes (F/B) ratio and increased the abundance of Proteobacteria and Episilonbacteria. And mAPS significantly decreased the expression of colonic G-protein-coupled receptors (GPR) 41 and 43, but it had little effect on the profile of fecal short-chain fatty acids (SCFAs) ( Zhong et al., 2022 ). Additionally, Liu et al. demonstrated that APS significantly regulated gut microbial dysbiosis while also recovering the abnormality of fecal metabolism, including glycolysis/gluconeogenesis metabolism and pyruvate metabolism ( Liu et al., 2022 ). Ying et al. aimed to evaluate the protective effect of Cordyceps sinensis polysaccharides (CSP). They found that CSP could increase the abundance of probiotics and decrease pathogenic bacteria. It reduced the side effects of cyclophosphamide (Cy) on intestinal mucosal immunity and gut microbiota ( Ying et al., 2020 ). Turmeric polysaccharides (TPS) were found to increase the abundance of probiotics, such as Lactobacillus and Clostridium-UCG-014, and exert their gut barrier functions through the activation of the aryl hydrocarbon receptor (AhR) to upregulate epithelial tight junction proteins ( Yang et al., 2021 ). Jing et al. used Codonopsis pilosula polysaccharides to treat colitis in model mice with Dextran Sulfate Sodium (DSS)-Induced, and they found that this medicine could stimulate the growth of important probiotics, inhibit the growth of pathogenic bacteria, and enhance the production of short-chain fatty acids ( Jing et al., 2018 ).
Table 1
The polysaccharides from TCM regulated gut microbiome.
↑ increase. ↓ decrease.
3.1.2. Saponin
Saponins, glycosides widely distributed in TCM, include a diverse group of compounds characterized by their structure, which contains a steroidal or triterpenoid aglycone and one or more sugar chains ( Güçlü-Üstündağ and Mazza, 2007 ). Which possess a multitude of biological activities such as antitumor activities, antimicrobial activity, antiviral activity, etc. ( Kimura et al., 2019 ; Sharma et al., 2021 ). In recent years, it has been found that the components of saponins from TCM play a role in disease treatment by regulating intestinal flora ( Table 2 ). Akebia saponin D (ASD) has been shown to treat hyperlipidemic rats induced by a high-fat diet by regulating the intestinal microbiota, and it could partially recover both metabolism dysfunction and the intestinal environment through several metabolic pathways and modulation of the microbial community ( Zhou et al., 2019 ). Guo et al. demonstrated that ginsenoside Rg1 possessed a neuroprotective effect on tree shrew model for Alzheimer’s disease, and may have a close association with the microbiota of the large intestine by significantly reducing the abundance of Bacteroidetes ( Guo et al., 2021b ). Alike, Chen et al. found that ginsenoside Rg1 could mitigate morphine dependence via regulation of gut microbiota and inhibit gut microbiota-derived tryptophan metabolism and reduce serotonin ( Chen et al., 2022a ). Astragalus has the effects of anti-tumor, lowering blood pressure, lowering blood sugar, and improving human immunity. Gong et al. found in animal experiments that Astragaloside IV plays a hypoglycemic role by regulating intestinal flora and AMPK/SIRT1 and PI3K/AKT pathways ( Gong et al., 2021 ). On the other hand, intestinal microorganisms can also affect the metabolism of saponins in vivo ( Dong et al., 2017 ; Zhang et al., 2018 ).
Table 2
The saponins from TCM regulated gut microbiome.
3.1.3. Flavonoids
Smilax china L ., commonly known as “Baqia” is not just a comestible; it was also used as traditional herbal medicine in China. It contains multifarious naturally bioactive compounds, such as flavonoids, polyphenols, and steroidal saponins ( Table 3 ). Li et al. investigated the effects of Smilax chinensis L . flavonoid (SCF) on obesity and changes in gut microbiota. Their results found that SCF modulated the composition of gut microbiota and decreased the production of SCFAs, resulting in reduced energy absorption and subsequent weight loss in the mice ( Li et al., 2021 ). The findings of Wang et al. provide evidence that Acanthopanax senticosus total flavonoids (ASTFs) have significant anti-inflammatory properties on LPS-induced intestinal inflammation, preserve the integrity of the intestinal barrier, and regulate gut microbiota homeostasis ( Wang X. et al., 2022 ). Baicalin has a variety of pharmacological effects, including anti-inflammation, anti-infection, anti-apoptosis, anti-allergy, and so on. Some scholars have found that baicalin rebalances the gut microbial composition pattern impaired by ionizing radiation (IR) and alleviates IR-induced apoptosis of the gut microbiota ( Wang et al., 2020b ). In addition, other scholars have also done research on baicalin; they found that baicalin can improve abnormal metabolism and gut microbiota in high-fat diet (HFD)-induced metabolic syndrome (MetS) in mice ( Lin et al., 2022 ). Glycosides and flavonoids from P. thomsonii leaves (PL) alleviated type 2 diabetes in high-fat diet plus streptozotocin-induced mice. This process may be associated with the biological activity that Glycosides and flavonoids from PL could increase intestinal probiotics to improve metabolic disorders caused by diabetes and decrease the level of Clostridium celatum to relieve inflammation ( Zhang et al., 2022 ).
Table 3
The flavonoids from TCM regulated gut microbiome.
3.1.4. Alkaloid
Alkaloid is an important natural organic compound that is one of the important effective components in Chinese herbal medicine. It has a variety of biological activities and pharmacological effects, mainly antibacterial, anti-inflammatory, liver protection, and nervous system effects. It has been confirmed that the components of alkaloids from TCM play a role in disease treatment by regulating gut microbiota ( Table 4 ). Zhang et al. confirmed that alkaloids from Sophora alopecuroides L. can improve depression in mice; this biological phenomenon is associated with modulating gut microbiota and gut microbiota directly participating in the metabolic process of the host ( Zhang et al., 2021a ). Berberine, the major active ingredient of the Chinese herb Coptis chinensis (Huang-Lian), has been used by clinicians to treat bacterial diarrhea. Zhang et al. found that berberine can alter the functions and metabolites of the gut microbiota. And Zhao et al. used berberine to treat a mouse model with acute graft-versus-host disease (aGVHD); their results suggest that the berberine could remodel gut microbiota and prevent colonic barrier impairment ( Zhao et al., 2022 ). Other scholars also found that berberine increased the abundance of beneficial bacteria that can produce SCFAs, and the SCFAs can directly alleviate the accumulation of uric acid ( Shan et al., 2022 ). Song et al. demonstrated that sinomenine can inhibit the inflammatory response by modulating gut microbiota and restoring the intestinal barrier via the aryl hydrocarbon receptor/Nrf2-dependent pathway ( Song et al., 2021 ).
Table 4
The alkaloid from TCM regulated gut microbiome.
3.2. Chinese herbal compound
Traditional Chinese medicine compound usually consists of two or more medicinal flavors, has relatively prescriptive processing methods and use methods, and is designed for relatively certain diseases and syndromes. At present, many compounds have been applied to the treatment of gut microbiota ( Table 5 ). For example, the combination of Astragalus membranaceus and Salvia miltiorrhiza (AS) is an effective prescription that is widely used to treat chronic kidney disease (CKD) clinically in traditional Chinese medicine. AS could alleviate renal fibrosis and metabolism through the “gut-kidney axis”, Han et al. found that AS restored the intestinal barrier and flora structure and increased butyric acid and lactic acid to exert the above effects ( Han et al., 2021 ). Zhao et al. also found that the Chinese herb FuZhengHuaYuJiangZhuTongLuoFang prescription (FZHY) can effectively treat CKD; the pathways included the regulation of gut microbiota ( Chen et al., 2022b ). Other researchers found that Chinese herbs have an anti-aging effect. Luo et al. revealed that FuFang Zhenshu TiaoZhi (FTZ) can moderately correct the aging process, which may be related to modulating the balance of intestinal microecology and restoring inflammation ( Luo et al., 2020 ). Painong Powder was developed by Zhongjing Zhang in the Han Dynasty and has been widely used to treat ulcerative colitis (UC) in China for thousands of years. The experimental research confirmed that Painong-San (PNS) extract alleviates colitis in mice by modulating gut microbiota and restoring intestinal barrier function ( Wang K. et al., 2022 ). Xuanbai Chengqi decoction (XBCQ) is a representative traditional Chinese medicine prescription used by Wu Jutong in the Qing Dynasty. It has been widely used to treat a variety of common respiratory diseases in China. There are also scholars studying his role in the regulation of intestinal flora. Wang et al. demonstrated that XBCQ could alleviate chronic obstructive pulmonary disease (COPD) exacerbations by reshaping the gut microbiota and improving the Th17/Treg balance ( Wang et al., 2021b ). And the studies have shown that Yangyin Qingfei decoction can effectively improve the skin damage of lung cancer patients after radiotherapy, help the growth and propagation of beneficial bacteria in the intestinal tract of lung cancer patients, and regulate the structural balance of intestinal flora ( Pan and Zhang 2023 ).
Table 5
Chinese herbal compound.
4. Effection of TCMs on prevention and treatment of NSCLC
4.1. active components of tcm.
Ganoderma lucidum polysaccharides (GLP) were derived from Ganoderma lucidum (lingzhi in Chinese). Gynostemma pentaphyllum saponins (GpS) are derived from Gynostemma pentaphyllum. Both are valuable traditional Chinese medicines. Khan et al. provide strong evidence that the cancer-preventive and therapeutic functions of GpS and GLP are through the dynamic modulation of GM and host immune responses. The specific performance is an improved inflamed gut barrier and promoted short-chain fatty acids (SCFAs) producing bacteria ( Khan et al., 2019 ). Saponins, a novel type of plant-derived secondary metabolites, modulate gut microbiota composition and exhibit anti-metastasis activities in multiple tumors, including lung adenocarcinoma, alcohol-induced liver disease, and colorectal cancer ( Khan et al., 2019 ; Chen et al., 2020a ; Zhou et al., 2021 ). Ginsenoside Rh2 (G-Rh2), a major bioactive ingredient in ginseng, suggested the therapeutic effects of G-Rh2 on lung cancer. G-Rh2 regulated the phenotype of macrophages and affected the migration of non-small cell lung cancer (NSCLC) cells ( Li et al., 2018 ). On the other hand, the gastrointestinal microbiome plays an important role in drug metabolism. Ginsenoside is difficult for the molecules to directly exert the pharmacological effects, and the gut flora and its metabolites can improve this process ( Yang et al., 2020 ). A triterpenoid saponin glycoside found in licorice roots is called glycyrrhizic acid (GA). Wu et al. discovered that glycyrrhizin prevented PDX mice from developing lung tumors ( Wu et al., 2018 ). And high amount of High Mobility Group Box 1 (HMGB1) facilitated lung cancer cell invasion and migration, which glycyrrhizin reduced. Here, Qiu et al. found that GA regulates GM to inhibit the establishment of pre-metastatic niches and metastasis that are promoted by HFD by reducing M1-like colonic macrophages via LPS/HMGB1/NF-κB signaling. The regulation of intestinal flora is mainly reflected in decreasing the Clostridiales order and Desulfovibrio genus, and reducing the ratio between Firmicutes and Bacteroidetes ( Qiu et al., 2019 ). A polysaccharide obtained from Spirulina (PSP) decreased the tumor volume and weight of the lung cancer-bearing mice through regulated arachidonic acid metabolism and the balance of gut microbiota. And the PSP increased the abundance of Lactobacillus , Allobaculum , Alloprevotella , and Olsenella and decreased Bacteroides and Acinetobacter ( Lu et al., 2022 ).
4.2. Compound formulation of TCM
Bovis calculus (Bos taurus domesticus Gmelin), Olibanum (Boswcllia bhaurdajiana Birdw., Boszvellia carterii Birdw.), Myrrha, and Moschus are the four herbs that make up the Xihuang pill (XHW), a Chinese medicine formula that has been approved (state medical permit number Z11020073) (Commiphora molmol Engl., Commiphora myrrha Engl.) ( Zhang et al., 2021b ). It has been widely used in the treatment of a variety of malignant tumors in clinics in China, including breast cancer, lung cancer, gastric cancer, and other malignant tumors ( Wang et al., 2020a ; Wang et al., 2021a ; Ge et al., 2022 ). In mice with Lewis lung cancer, Cao et al. investigated the anti-lung cancer impact of XHW coupled with anlotinib (LLC). They clarified the regulatory features of XHW in enhancing anlotinib’s anti-lung cancer impact using GM and transcriptomics. The outcomes demonstrated that LLC-bearing mice receiving a combination therapy of XHW and anlotinib effectively suppressed tumor growth. Additionally, XHW’s impact on the control of gut microbiota was significant, as revealed by 16s rRNA sequencing research. The percentage of the helpful bacteria Bacteroides and g_norank_f_ Muribaculaceae increased as a result ( Cao et al., 2022 ). BuFeiXiaoJiYin (BFXJY) is a traditional Chinese medicine (TCM) compound that has been shown to have good effects in the treatment of lung cancer by ameliorating the NLRP3 inflammation response and regulating gut microbiota. Jiang et al. found that BFXJY reduced the relative abundance of Firmicutes and Verrucomicrobia , increased the relative abundance of Bacteroidetes and Epsilonbacteraeota , and enhanced the relative abundance of Deferribacteres ( Jiang et al., 2022 ). A Chinese clinical study suggests that Huayu Kangai decoction enema is effective in the treatment of lung cancer and can improve the level of lactic acid bacteria and bifidobacteria ( Jia et al., 2023 ).
In sum, many of the herbal medicines could modulate the relationship between the host and the gut microbiota (GM) to exert their beneficial properties on the host. However, most current studies are mainly one-way studies, which mainly observe the changes in flora and metabolites after the treatment of diseases with traditional Chinese medicine. There will be some problems in this process. For example, Chinese herbs themselves will affect intestinal flora, and intestinal flora is needed for the metabolism and absorption of Chinese herbs. Changes in flora caused by these life processes may not play a major role in the treatment of diseases. In addition, the metabolic components of traditional Chinese medicine are diverse, and some metabolites may be repeated with those of the bacterial community. Therefore, there are many problems in the study of the treatment of lung cancer by regulating intestinal flora with traditional Chinese medicine, and researchers could try their best to improve the experimental scheme and reduce the above problems.
5. Conclusion
There are objective differences between lung cancer patients and healthy people in intestinal flora, and more and more studies are gradually proving that by regulating intestinal flora, increasing probiotics, and reducing harmful bacteria content, the cancer inhibitory signaling pathway can be more activated, anti-tumor immunity can be enhanced, thus inducing apoptosis of cancer cells or preventing recurrence and metastasis, and it can play a synergistic role with tumor therapeutic drugs. This paper summarizes and analyzes the regulation of intestinal flora by active ingredients of TCM and Chinese herbal compounds and their intervention effects. Various microbial-related therapeutic means represented by fecal bacteria transplantation and Chinese medicine intervention have been widely used in the fields of diabetes, kidney disease, liver disease, inflammatory bowel disease, and are expected to be popularized in the treatment of cancer, psychiatric diseases, cardiovascular diseases, and so on. Which is conducive to providing new strategies and ideas for clinical prevention and treatment of NSCLC. At present, studies show that TCM regulates intestinal flora to repair the intestinal mucosal barrier and treats NSCLC mainly in two aspects: by affecting immune cells to restore normal immune function of the intestinal mucosa and by improving the sensitivity of antitumor drugs by regulating intestinal flora itself or metabolites. On the other hand, some TCM ingredients cannot be directly absorbed into the blood to play a role in the treatment of diseases, and intestinal flora can achieve drug transformation in this process. With the advantages of multi-target, all-directional, multi-component, and light side effects, TCM intervention in NSCLC has been recognized as a broad prospect. However, the composition of traditional Chinese medicine is complex, and its mechanism of action has not been fully elucidated. In addition, there are a few studies on the treatment of NSCLC through the regulation of intestinal flora by traditional Chinese medicine. Therefore, a large number of basic and clinical studies are needed in the TCM treatment of NSCLC.
Author contributions
XW and LH collected the articles, MC and JL made the figures. JX guided XW and LH in collecting articles and making figures. MW prerevised the manuscript. XW and LH wrote the manuscript and made the tables. XW and LH have the same contribution to the article. All authors contributed to the article and approved the submitted version.
Funding Statement
This work was supported by the Natural Science Foundation of China (NSFC:21272214).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
- Adak A., Khan M. R. (2019). An insight into gut microbiota and its functionalities . Cell. Mol. Life Sci. 76 , 473–493. doi: 10.1007/s00018-018-2943-4 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Berding K., Vlckova K., Marx W., Schellekens H., Stanton C., Clarke G., et al.. (2021). Diet and the microbiota–Gut–Brain axis: Sowing the seeds of good mental health . Adv. Nutr. 12 , 1239–1285. doi: 10.1093/advances/nmaa181 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Botticelli A., Vernocchi P., Marini F., Quagliariello A., Cerbelli B., Reddel S., et al.. (2020). Gut metabolomics profiling of non-small cell lung cancer (NSCLC) patients under immunotherapy treatment . J. Transl. Med. 18 , 49. doi: 10.1186/s12967-020-02231-0 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Cao B., Wang S., Li R., Wang Z., Li T., Zhang Y., et al.. (2022). Xihuang pill enhances anticancer effect of anlotinib by regulating gut microbiota composition and tumor angiogenesis pathway . Biomed Pharmacother 151 , 113081. doi: 10.1016/j.biopha.2022.113081 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Chen L., Chen M.-Y., Shao L., Zhang W., Rao T., Zhou H.-H., et al.. (2020. a). Panax notoginseng saponins prevent colitis-associated colorectal cancer development: the role of gut microbiota . Chin. J. Natural Medicines 18 , 500–507. doi: 10.1016/S1875-5364(20)30060-1 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Chen M., Jiang W., Xiao C., Yang W., Qin Q., Mao A., et al.. (2020. b). Sodium butyrate combined with docetaxel for the treatment of lung adenocarcinoma A549 cells by targeting Gli1 . OTT Volume 13 , 8861–8875. doi: 10.2147/OTT.S252323 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Chen Z., Lin Y., Zhou Q., Xiao S., Li C., Lin R., et al.. (2022. a). Ginsenoside Rg1 mitigates morphine dependence via regulation of gut microbiota, tryptophan metabolism, and serotonergic system function . Biomed Pharmacother 150 , 112935. doi: 10.1016/j.biopha.2022.112935 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Chen Z., Qian X., Chen S., Fu X., Ma G., Zhang A. (2020. c). Akkermansia muciniphila enhances the antitumor effect of cisplatin in Lewis lung cancer mice . J. Immunol. Res. 2020 , 1–13. doi: 10.1155/2020/2969287 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Chen Z., Wu S., Zeng Y., Chen Z., Li X., Li J., et al.. (2022. b). FuZhengHuaYuJiangZhuTongLuoFang prescription modulates gut microbiota and gut-derived metabolites in UUO rats . Front. Cell. Infect. Microbiol. 12 . doi: 10.3389/fcimb.2022.837205 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Chen Y., Zhou J., Wang L. (2021. b). Role and mechanism of gut microbiota in human disease . Front. Cell. Infect. Microbiol. 11 . doi: 10.3389/fcimb.2021.625913 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Chen L., Zhou X., Wang Y., Wang D., Ke Y., Zeng X. (2021. a). Propionate and butyrate produced by gut microbiota after probiotic supplementation attenuate lung metastasis of melanoma cells in mice . Mol. Nutr. Food Res. 65 , 2100096. doi: 10.1002/mnfr.202100096 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Corrêa R. O., Castro P. R., Moser R., Ferreira C. M., Quesniaux V. F. J., Vinolo M. A. R., et al.. (2022). Butyrate: Connecting the gut-lung axis to the management of pulmonary disorders . Front. Nutr. 9 . doi: 10.3389/fnut.2022.1011732 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Cristiano C., Cuozzo M., Coretti L., Liguori F. M., Cimmino F., Turco L., et al.. (2022). Oral sodium butyrate supplementation ameliorates paclitaxel-induced behavioral and intestinal dysfunction . Biomed Pharmacother 153 , 113528. doi: 10.1016/j.biopha.2022.113528 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Ding R., Goh W.-R., Wu R., Yue X., Luo X., Khine W. W. T., et al.. (2019). Revisit gut microbiota and its impact on human health and disease . J. Food Drug Anal. 27 , 623–631. doi: 10.1016/j.jfda.2018.12.012 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Dong W.-W., Xuan F.-L., Zhong F.-L., Jiang J., Wu S., Li D., et al.. (2017). Comparative analysis of the rats’ gut microbiota composition in animals with different ginsenosides metabolizing activity . J. Agric. Food Chem. 65 , 327–337. doi: 10.1021/acs.jafc.6b04848 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Fan Y., Pedersen O. (2021). Gut microbiota in human metabolic health and disease . Nat. Rev. Microbiol. 19 , 55–71. doi: 10.1038/s41579-020-0433-9 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Fulde M., Hornef M. W. (2014). Maturation of the enteric mucosal innate immune system during the postnatal period . Immunol. Rev. 260 , 21–34. doi: 10.1111/imr.12190 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Ge A., Yang K., Deng X., Zhao D., Ge J., Liu L. (2022). The efficacy and safety of xihuang pill/capsule in adjuvant treatment of breast cancer: A systematic review and meta-analysis of 26 randomized controlled trials . J. Ethnopharmacol 295 , 115357. doi: 10.1016/j.jep.2022.115357 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Georgiou K., Marinov B., Farooqi A. A., Gazouli M. (2021). Gut microbiota in lung cancer: Where do we stand ? IJMS 22 , 10429. doi: 10.3390/ijms221910429 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Gong P., Xiao X., Wang S., Shi F., Liu N., Chen X., et al.. (2021). Hypoglycemic effect of astragaloside IV via modulating gut microbiota and regulating AMPK/SIRT1 and PI3K/AKT pathway . J. Ethnopharmacol 281 , 114558. doi: 10.1016/j.jep.2021.114558 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Grenda A., Iwan E., Chmielewska I., Krawczyk P., Giza A., Bomba A., et al.. (2022). Presence of akkermansiaceae in gut microbiome and immunotherapy effectiveness in patients with advanced non-small cell lung cancer . AMB Expr 12 , 86. doi: 10.1186/s13568-022-01428-4 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Güçlü-Üstündağ Ö., Mazza G. (2007). Saponins: Properties, applications and processing . Crit. Rev. Food Sci. Nutr. 47 , 231–258. doi: 10.1080/10408390600698197 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Guo C., Guo D., Fang L., Sang T., Wu J., Guo C., et al.. (2021. a). Ganoderma lucidum polysaccharide modulates gut microbiota and immune cell function to inhibit inflammation and tumorigenesis in colon . Carbohydr. Polymers 267 , 118231. doi: 10.1016/j.carbpol.2021.118231 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Guo Y., Wang L., Lu J., Jiao J., Yang Y., Zhao H., et al.. (2021. b). Ginsenoside Rg1 improves cognitive capability and affects the microbiota of large intestine of tree shrew model for alzheimer’s disease . Mol. Med. Rep. 23 , 291. doi: 10.3892/mmr.2021.11931 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Hakozaki T., Nolin-Lapalme A., Kogawa M., Okuma Y., Nakamura S., Moreau-Amaru D., et al.. (2022). Cancer cachexia among patients with advanced non-Small-Cell lung cancer on immunotherapy: An observational study with exploratory gut microbiota analysis . Cancers 14 , 5405. doi: 10.3390/cancers14215405 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Hakozaki T., Richard C., Elkrief A., Hosomi Y., Benlaïfaoui M., Mimpen I., et al.. (2020). The gut microbiome associates with immune checkpoint inhibition outcomes in patients with advanced non–small cell lung cancer . Cancer Immunol. Res. 8 , 1243–1250. doi: 10.1158/2326-6066.CIR-20-0196 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Han C., Jiang Y., Li W., Liu Y. (2021). Astragalus membranaceus and salvia miltiorrhiza ameliorates cyclosporin a-induced chronic nephrotoxicity through the “gut-kidney axis” . J. Ethnopharmacol 269 , 113768. doi: 10.1016/j.jep.2020.113768 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Heintz-Buschart A., Wilmes P. (2018). Human gut microbiome: Function matters . Trends Microbiol. 26 , 563–574. doi: 10.1016/j.tim.2017.11.002 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Hooper L. V., Littman D. R., Macpherson A. J. (2012). Interactions between the microbiota and the immune system . Science 336 , 1268–1273. doi: 10.1126/science.1223490 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Huang J., Liu D., Wang Y., Liu L., Li J., Yuan J., et al.. (2022). Ginseng polysaccharides alter the gut microbiota and kynurenine/tryptophan ratio, potentiating the antitumour effect of antiprogrammed cell death 1/programmed cell death ligand 1 (anti-PD-1/PD-L1) immunotherapy . Gut 71 , 734–745. doi: 10.1136/gutjnl-2020-321031 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Hufnagl K., Pali-Schöll I., Roth-Walter F., Jensen-Jarolim E. (2020). Dysbiosis of the gut and lung microbiome has a role in asthma . Semin. Immunopathol. 42 , 75–93. doi: 10.1007/s00281-019-00775-y [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Jia X., Liu Q., Zhao K. (2023). Effect of huayu anticancer decoction enema combined with auricular point pressing on intestinal flora and inflammatory factors in lung cancer patients . Minerva Surg. 78 , 123–125. doi: 10.23736/S2724-5691.21.09339-4 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Jiang R., Wang T., Lan Q., Qin Y., Man T., Sun H., et al.. (2022). BuFeiXiaoJiYin ameliorates the NLRP3 inflammation response and gut microbiota in mice with lung cancer companied with qi-yin deficiency . Cancer Cell Int. 22 , 121. doi: 10.1186/s12935-022-02543-9 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Jin Y., Dong H., Xia L., Yang Y., Zhu Y., Shen Y., et al.. (2019). The diversity of gut microbiome is associated with favorable responses to anti–programmed death 1 immunotherapy in Chinese patients with NSCLC . J. Thorac. Oncol. 14 , 1378–1389. doi: 10.1016/j.jtho.2019.04.007 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Jing Y., Li A., Liu Z., Yang P., Wei J., Chen X., et al.. (2018). Absorption of Codonopsis pilosula saponins by coexisting polysaccharides alleviates gut microbial dysbiosis with dextran sulfate sodium-induced colitis in model mice . BioMed. Res. Int. 2018 , 1–18. doi: 10.1155/2018/1781036 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Kamada N., Chen G. Y., Inohara N., Núñez G. (2013). Control of pathogens and pathobionts by the gut microbiota . Nat. Immunol. 14 , 685–690. doi: 10.1038/ni.2608 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Khan I., Huang G., Li X., Liao W., Leong W. K., Xia W., et al.. (2019). Mushroom polysaccharides and jiaogulan saponins exert cancer preventive effects by shaping the gut microbiota and microenvironment in apc mice . Pharmacol. Res. 148 , 104448. doi: 10.1016/j.phrs.2019.104448 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Kim K., Kwon O., Ryu T., Jung C., Kim J., Min J., et al.. (2019). Propionate of a microbiota metabolite induces cell apoptosis and cell cycle arrest in lung cancer . Mol. Med. Rep . 20 , 1569–1574. doi: 10.3892/mmr.2019.10431 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Kimura M., Sasaki K., Fukutani Y., Yoshida H., Ohsawa I., Yohda M., et al.. (2019). Anticancer saponin OSW-1 is a novel class of selective golgi stress inducer . Bioorganic Medicinal Chem. Lett. 29 , 1732–1736. doi: 10.1016/j.bmcl.2019.05.022 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Knezevic J., Starchl C., Tmava Berisha A., Amrein K. (2020). Thyroid-Gut-Axis: How does the microbiota influence thyroid function ? Nutrients 12 , 1769. doi: 10.3390/nu12061769 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Leigh S.-J., Morris M. J. (2020). Diet, inflammation and the gut microbiome: Mechanisms for obesity-associated cognitive impairment . Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 1866 , 165767. doi: 10.1016/j.bbadis.2020.165767 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Li H., Huang N., Zhu W., Wu J., Yang X., Teng W., et al.. (2018). Modulation the crosstalk between tumor-associated macrophages and non-small cell lung cancer to inhibit tumor migration and invasion by ginsenoside Rh2 . BMC Cancer 18 , 579. doi: 10.1186/s12885-018-4299-4 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Li X., Yang L., Li J., Lin L., Zheng G. (2021). A flavonoid-rich Smilax china l. extract prevents obesity by upregulating the adiponectin-receptor/AMPK signalling pathway and modulating the gut microbiota in mice . Food Funct. 12 , 5862–5875. doi: 10.1039/D1FO00282A [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Lianqun J., Xing J., Yixin M., Si C., Xiaoming L., Nan S., et al.et al. (2021). Comprehensive multiomics analysis of the effect of ginsenoside Rb1 on hyperlipidemia . Aging 13 , 9732–9747. doi: 10.18632/aging.202728 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Lin Y., Wang Z.-Y., Wang M.-J., Jiang Z.-M., Qin Y.-Q., Huang T.-Q., et al.. (2022). Baicalin attenuate diet-induced metabolic syndrome by improving abnormal metabolism and gut microbiota . Eur. J. Pharmacol. 925 , 174996. doi: 10.1016/j.ejphar.2022.174996 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Liu X., Li M., Jian C., Wei F., Liu H., Li K., et al.. (2022). Astragalus polysaccharide alleviates constipation in the elderly Via modification of gut microbiota and fecal metabolism . Rejuvenation Res. 25 , 275–290. doi: 10.1089/rej.2022.0039 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Lu Y., Peng B., Lin Y., Lin Q., Xia X., Zhong S., et al.. (2022). Spirulina polysaccharide induces the metabolic shifts and gut microbiota change of lung cancer in mice . Curr. Res. Food Sci. 5 , 1313–1319. doi: 10.1016/j.crfs.2022.08.010 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Luo D., Chen K., Li J., Fang Z., Pang H., Yin Y., et al.. (2020). Gut microbiota combined with metabolomics reveals the metabolic profile of the normal aging process and the anti-aging effect of FuFang zhenshu TiaoZhi(FTZ) in mice . Biomed Pharmacother 121 , 109550. doi: 10.1016/j.biopha.2019.109550 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Luo S., Zhang X., Huang S., Feng X., Zhang X., Xiang D. (2022). A monomeric polysaccharide from polygonatum sibiricum improves cognitive functions in a model of alzheimer’s disease by reshaping the gut microbiota . Int. J. Biol. Macromolecules 213 , 404–415. doi: 10.1016/j.ijbiomac.2022.05.185 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Mirzaei R., Afaghi A., Babakhani S., Sohrabi M. R., Hosseini-Fard S. R., Babolhavaeji K., et al.. (2021). Role of microbiota-derived short-chain fatty acids in cancer development and prevention . Biomed Pharmacother 139 , 111619. doi: 10.1016/j.biopha.2021.111619 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Nagasaka M., Sexton R., Alhasan R., Rahman S., Azmi A. S., Sukari A. (2020). Gut microbiome and response to checkpoint inhibitors in non-small cell lung cancer–a review . Crit. Rev. Oncol/Hematol 145 , 102841. doi: 10.1016/j.critrevonc.2019.102841 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Ni L., Kong L., Tang Y., Nie Y., Wang X., Yang X., et al.. (2022). The experimental exploration of TCM theory “Treating the same disease with different approaches” on an ulcerative colitis model . Evidence-Based Complementary Altern. Med. 2022 , 1–11. doi: 10.1155/2022/4916540 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Pan T. T., Zhang A. Q. (2023). Effect of yangyin qingfei decoction on skin damage and intestinal flora in patients with lung cancer after radiotherapy . Liaoning J. Traditional Chin. Med. 01 , 97–100. doi: 10.13192/j.iSSN.1000-1719.2023.01.028 [ CrossRef ] [ Google Scholar ]
- Peng Z., Cheng S., Kou Y., Wang Z., Jin R., Hu H., et al.. (2020). The gut microbiome is associated with clinical response to anti–PD-1/PD-L1 immunotherapy in gastrointestinal cancer . Cancer Immunol. Res. 8 , 1251–1261. doi: 10.1158/2326-6066.CIR-19-1014 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Pizzo F., Maroccia Z., Hammarberg Ferri I., Fiorentini C. (2022). Role of the microbiota in lung cancer: Insights on prevention and treatment . IJMS 23 , 6138. doi: 10.3390/ijms23116138 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Qiu M., Huang K., Liu Y., Yang Y., Tang H., Liu X., et al.. (2019). Modulation of intestinal microbiota by glycyrrhizic acid prevents high-fat diet-enhanced pre-metastatic niche formation and metastasis . Mucosal Immunol. 12 , 945–957. doi: 10.1038/s41385-019-0144-6 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Routy B., Le Chatelier E., Derosa L., Duong C. P. M., Alou M. T., Daillère R., et al.. (2018). Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors . Science 359 , 91–97. doi: 10.1126/science.aan3706 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Rutsch A., Kantsjö J. B., Ronchi F. (2020). The gut-brain axis: How microbiota and host inflammasome influence brain physiology and pathology . Front. Immunol. 11 . doi: 10.3389/fimmu.2020.604179 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Sánchez-Alcoholado L., Ordóñez R., Otero A., Plaza-Andrade I., Laborda-Illanes A., Medina J. A., et al.. (2020). Gut microbiota-mediated inflammation and gut permeability in patients with obesity and colorectal cancer . IJMS 21 , 6782. doi: 10.3390/ijms21186782 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Shan B., Wu M., Chen T., Tang W., Li P., Chen J. (2022). Berberine attenuates hyperuricemia by regulating urate transporters and gut microbiota . Am. J. Chin. Med. 50 , 2199–2221. doi: 10.1142/S0192415X22500951 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Sharma P., Tyagi A., Bhansali P., Pareek S., Singh V., Ilyas A., et al.. (2021). Saponins: Extraction, bio-medicinal properties and way forward to anti-viral representatives . Food Chem. Toxicol. 150 , 112075. doi: 10.1016/j.fct.2021.112075 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Song W., Yang X., Wang W., Wang Z., Wu J., Huang F. (2021). Sinomenine ameliorates septic acute lung injury in mice by modulating gut homeostasis via aryl hydrocarbon receptor/Nrf2 pathway . Eur. J. Pharmacol. 912 , 174581. doi: 10.1016/j.ejphar.2021.174581 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Sun S.-S., Wang K., Ma K., Bao L., Liu H.-W. (2019). An insoluble polysaccharide from the sclerotium of poria cocos improves hyperglycemia, hyperlipidemia and hepatic steatosis in ob/ob mice via modulation of gut microbiota . Chin. J. Natural Medicines 17 , 3–14. doi: 10.1016/S1875-5364(19)30003-2 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Takada K., Shimokawa M., Takamori S., Shimamatsu S., Hirai F., Tagawa T., et al.. (2021). Clinical impact of probiotics on the efficacy of anti-PD -1 monotherapy in patients with nonsmall cell lung cancer: A multicenter retrospective survival analysis study with inverse probability of treatment weighting . Intl J. Cancer 149 , 473–482. doi: 10.1002/ijc.33557 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Tang W. H. W., Li D. Y., Hazen S. L. (2019). Dietary metabolism, the gut microbiome, and heart failure . Nat. Rev. Cardiol. 16 , 137–154. doi: 10.1038/s41569-018-0108-7 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Tang Q., Liu R., Chu G., Wang Y., Cui H., Zhang T., et al.. (2022). A comprehensive analysis of microflora and metabolites in the development of ulcerative colitis into colorectal cancer based on the lung–gut correlation theory . Molecules 27 , 5838. doi: 10.3390/molecules27185838 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Vernocchi P., Gili T., Conte F., Del Chierico F., Conta G., Miccheli A., et al.. (2020). Network analysis of gut microbiome and metabolome to discover microbiota-linked biomarkers in patients affected by non-small cell lung cancer . IJMS 21 , 8730. doi: 10.3390/ijms21228730 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Walker R. L., Vlamakis H., Lee J. W. J., Besse L. A., Xanthakis V., Vasan R. S., et al.. (2021). Population study of the gut microbiome: associations with diet, lifestyle, and cardiometabolic disease . Genome Med. 13 , 188. doi: 10.1186/s13073-021-01007-5 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wang M., Dong Y., Wu J., Li H., Zhang Y., Fan S., et al.. (2020. b). Baicalein ameliorates ionizing radiation-induced injuries by rebalancing gut microbiota and inhibiting apoptosis . Life Sci. 261 , 118463. doi: 10.1016/j.lfs.2020.118463 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wang J., Hou D., Peng Y., Xiong J., Xiong L. (2020. a). Efficacy and safety of xihuang pill for lung cancer: A protocol for systematic review and meta-analysis . Medicine 99 , e22516. doi: 10.1097/MD.0000000000022516 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wang J., Hou D., Peng Y., Xiong J., Xiong L., Tan X. (2021. a). Efficacy and safety of xihuang pill for gastric cancer: A protocol for systematic review and meta-analysis . Medicine 100 , e25726. doi: 10.1097/MD.0000000000025726 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wang Y., Li N., Li Q., Liu Z., Li Y., Kong J., et al.. (2021. b). Xuanbai chengqi decoction ameliorates pulmonary inflammation via reshaping gut microbiota and rectifying Th17/Treg imbalance in a murine model of chronic obstructive pulmonary disease . COPD Volume 16 , 3317–3335. doi: 10.2147/COPD.S337181 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wang Y., Wei J., Zhang W., Doherty M., Zhang Y., Xie H., et al.. (2022. a). Gut dysbiosis in rheumatic diseases: A systematic review and meta-analysis of 92 observational studies . eBioMedicine 80 , 104055. doi: 10.1016/j.ebiom.2022.104055 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wang X., Zhang X., Su J., Chu X. (2022. b). Acanthopanax senticosus total flavonoids alleviate lipopolysaccharide-induced intestinal inflammation and modulate the gut microbiota in mice . Bioscience Rep. 42 , BSR20212670. doi: 10.1042/BSR20212670 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Weersma R. K., Zhernakova A., Fu J. (2020). Interaction between drugs and the gut microbiome . Gut 69 , 1510–1519. doi: 10.1136/gutjnl-2019-320204 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wu X., Wang W., Chen Y., Liu X., Wang J., Qin X., et al.. (2018). Glycyrrhizin suppresses the growth of human NSCLC cell line HCC827 by downregulating HMGB1 level . BioMed. Res. Int. 2018 , 1–7. doi: 10.1155/2018/6916797 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Xiao X., Xu Y., Chen H. (2020). Sodium butyrate-activated TRAF6-TXNIP pathway affects A549 cells proliferation and migration . Cancer Med. 9 , 3477–3488. doi: 10.1002/cam4.2564 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Yang C., Du Y., Ren D., Yang X., Zhao Y. (2021). Gut microbiota-dependent catabolites of tryptophan play a predominant role in the protective effects of turmeric polysaccharides against DSS-induced ulcerative colitis . Food Funct. 12 , 9793–9807. doi: 10.1039/D1FO01468D [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Yang L., Zou H., Gao Y., Luo J., Xie X., Meng W., et al.. (2020). Insights into gastrointestinal microbiota-generated ginsenoside metabolites and their bioactivities . Drug Metab. Rev. 52 , 125–138. doi: 10.1080/03602532.2020.1714645 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Ying M., Yu Q., Zheng B., Wang H., Wang J., Chen S., et al.. (2020). Cultured cordyceps sinensis polysaccharides modulate intestinal mucosal immunity and gut microbiota in cyclophosphamide-treated mice . Carbohydr. Polymers 235 , 115957. doi: 10.1016/j.carbpol.2020.115957 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Yoon Y., Hrusch C. L., Fei N., Barrón G. M., Mills K. A. M., Hollinger M. K., et al.. (2022). Gut microbiota modulates bleomycin-induced acute lung injury response in mice . Respir. Res. 23 , 337. doi: 10.1186/s12931-022-02264-7 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Yu Y., Shen M., Song Q., Xie J. (2018). Biological activities and pharmaceutical applications of polysaccharide from natural resources: A review . Carbohydr. Polymers 183 , 91–101. doi: 10.1016/j.carbpol.2017.12.009 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zhang L., Li F., Qin W.-J., Fu C., Zhang X.-L. (2018). Changes in intestinal microbiota affect metabolism of ginsenoside re . Biomed. Chromatogr. 32 , e4284. doi: 10.1002/bmc.4284 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zhang M., Li A., Yang Q., Li J., Wang L., Liu X., et al.. (2021. a). Beneficial effect of alkaloids from sophora alopecuroides l. @ on CUMS-induced depression model mice via modulating gut microbiota . Front. Cell. Infect. Microbiol. 11 . doi: 10.3389/fcimb.2021.665159 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zhang Y.-Z., Yang J.-Y., Wu R.-X., Fang C., Lu H., Li H.-C., et al.. (2021. b). Network pharmacology–based identification of key mechanisms of xihuang pill in the treatment of triple-negative breast cancer stem cells . Front. Pharmacol. 12 . doi: 10.3389/fphar.2021.714628 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zhang S.-S., Zhang N.-N., Guo S., Liu S.-J., Hou Y.-F., Li S., et al.. (2022). Glycosides and flavonoids from the extract of Pueraria thomsonii benth leaf alleviate type 2 diabetes in high-fat diet plus streptozotocin-induced mice by modulating the gut microbiota . Food Funct. 13 , 3931–3945. doi: 10.1039/D1FO04170C [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zhao F., An R., Wang L., Shan J., Wang X. (2021). Specific gut microbiome and serum metabolome changes in lung cancer patients . Front. Cell. Infect. Microbiol. 11 . doi: 10.3389/fcimb.2021.725284 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zhao Y., Huang J., Li T., Zhang S., Wen C., Wang L. (2022). Berberine ameliorates aGVHD by gut microbiota remodelling, TLR4 signalling suppression and colonic barrier repairment for NLRP3 inflammasome inhibition . J. Cell. Mol. Medi 26 , 1060–1070. doi: 10.1111/jcmm.17158 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zhong M., Yan Y., Yuan H., Rong A., Xu G., Cai F., et al.. (2022). Astragalus mongholicus polysaccharides ameliorate hepatic lipid accumulation and inflammation as well as modulate gut microbiota in NAFLD rats . Food Funct. 13 , 7287–7301. doi: 10.1039/D2FO01009G [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zhou P., Yang X., Yang Z., Huang W., Kou J., Li F. (2019). Akebia saponin d regulates the metabolome and intestinal microbiota in high fat diet-induced hyperlipidemic rats . Molecules 24 , 1268. doi: 10.3390/molecules24071268 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zhou J., Zhang N., Zhao L., Wu W., Zhang L., Zhou F., et al.. (2021). Astragalus polysaccharides and saponins alleviate liver injury and regulate gut microbiota in alcohol liver disease mice . Foods 10 , 2688. doi: 10.3390/foods10112688 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Events Calendar
- Strauss Health Sciences Library
- Department A-Z Directory
- Campus Directory
- Faculty & Staff Resources
- Supporter & Alumni Resources
- Student Resources
- Mental Health Resources
- University Policies
- CU Anschutz Medical Campus
- CU Colorado Springs
- School of Dental Medicine
- Graduate School
School of Medicine
- College of Nursing
- Skaggs School of Pharmacy and Pharmaceutical Sciences
- Colorado School of Public Health
University of Colorado Cancer Center
- About Us Director's Message CU Cancer Center Leadership CU Cancer Center Staff National Cancer Institute-Designated Comprehensive Cancer Center National Comprehensive Cancer Network Partnerships Staff and Member Resources Contact Us Make a Gift
- Patients & Families Cancers We Treat Cancer Screening Multidisciplinary Clinics Second Opinions Supportive Care Survivorship
- Clinical Trials CCTO Team Women's Cancer Developmental Therapeutic Program Data Safety Monitoring Committee Protocol Review and Monitoring System Investigator-Initiated Trials Oncology Clinical Research Support Team
- Research Shared Resources Research Programs ORIEN Members Thoracic Oncology Research Initiative Head and Neck Cancer SPORE
- Education K-12 Training & Education Opportunities Undergraduate & Post-Baccalaureate Training & Education Opportunities Graduate, Post-Doctoral, & Medical Student Training & Education Opportunities Junior Faculty Professionals Training Grants Events Calendar
- Community Office of Community Outreach and Engagement Regional Network of Care

Home News Rosemary Rochford, PhD, Receives Henle Award for Decades of Research on Epstein-Barr Virus-Related Cancer back to News
Rosemary Rochford, PhD, Receives Henle Award for Decades of Research on Epstein-Barr Virus-Related Cancer
Rochford focuses on ebv- and malaria-related burkitt lymphoma in sub-saharan africa..
minute read
The malaria connection
Cultivating relationships.
Topics: Research , Awards , lymphoma

Rosemary Rochford, PhD
Related Stories
Research Awards lymphoma
Research Clinical Trials
CU Cancer Center Helps Lead National Trial to Evaluate Multicancer Blood Tests
Research Lung Cancer Medical Oncology
A Combination of Targeted Therapies Proves Effective Against Mutation-Driven Lung Cancer
Cancer center (som), cu anschutz, anschutz cancer pavilion.
1665 North Aurora Court
Aurora, CO 80045
720-848-0300
School of Medicine Home
Find a Doctor
Find a Researcher
Departments
School Profiles
Affiliate/Partner Hospitals
CU Medicine
Map and Parking
Health Sciences Library
Student Life
Colorado Springs Branch
- Website Feedback
- Privacy Policy
- Terms of Use
- Accessibility
- Accreditation
© 2024 The Regents of the University of Colorado , a body corporate. All rights reserved.
Accredited by the Higher Learning Commission . All trademarks are registered property of the University. Used by permission only.

IMAGES
VIDEO
COMMENTS
my thesis and providing help during this period whenever needed. ... my PhD and for the support throughout this thesis. Stephanie Kehlet September 2018 . 2 Preface The work presented in this thesis was carried out as a collaboration b etween The Technical University of ... relevance in lung cancer - implications of a new collagen chaperone ...
PhD thesis to obtain the degree of PhD at the University of Groningen on the authority of the Rector Magnificus Prof. E. Sterken and in accordance with ... Lung cancer is a result of multiple genetic alterations that accumulate during life21. In 1982, a human gene with transforming activity was identified in a lung ...
PhD thesis to obtain the degree of PhD at the University of Groningen on the authority of the Rector Magnificus Prof. C. Wijmenga and in accordance with ... Lung cancer is one of the most common cancers, and is a leading cause of cancer mortality among both men and women. Following the high number of new cases in prostate cancer in
Identification of lung cancer patients at higher risk for brain metastasis using microfluidics. University of Plymouth Peninsula Medical School. Applications are invited for a three-year PhD studentship. The studentship will start on 1st October 2024. . Project Description.
Lung cancer remains a devastating diagnosis to receive. The goal of this thesis was to better understand the impact of lung cancer and study how to integrate palliative and supportive care.
The U.S.-based National Lung Cancer Screening Trial (NLST) was launched in 2002 as a control-armed, prospective trial with the ultimate goal to determine whether LDCT screening has a beneficial effect in reducing lung cancer mortality. Between 2002 and 2004, 53,454 individuals with a calculated high risk for lung cancer (55-74 years of
Lung cancer mutations were successfully detected in small, formalin-fixed, paraffin embedded (FFPE) tumour tissue samples, and ctDNA, from lung cancer patients, using the same NGS technique, with a commercially available, targeted 50 gene cancer hotspot panel. Results are compared to a custom 22-gene panel.
Bradley, Stephen (2022) The role of imaging in the diagnosis of lung cancer in primary care. PhD thesis, University of Leeds. Abstract. Background Lung cancer is the leading cause of cancer death worldwide. The UK relies more heavily upon chest x-ray than many other high income countries. Little is known about the performance of the test, the ...
This dissertation deals with the development of a CAD system for lung cancer diagnosis, which is the second most common cancer in men after prostate cancer and in women after breast cancer. Moreover, lung cancer is considered the leading cause of cancer death among both genders in USA. Recently, the number of lung cancer patients has increased ...
approximately 2.09 million new cases and 1.76 million lung cancer-related deaths [2]. Lung cancer cases and deaths have increased significantly globally [2]. Approximately 85-88% of lung cancer cases are non-small cell lung carcinoma (NSCLS), and about 12-15% of lung cancer cases are small cell lung cancer (SCLC) [3]. Early lung cancer ...
2015. Kristie Hilliard The functional significance of the lung-liver axis during pneumonia Mentor: Lee J. Quinton Microbiology PhD. Rebecca Kusko Integrative transcriptomics in smoking related lung diseases Mentor: Avrum Spira Genetics & Genomics PhD. Teresa Wang Transcriptomics of the human airway epithelium reflect the physiologic response to ...
Lung cancer is a malignant tumor characterized by a rapid proliferation rate, less survivability, high mortality, and metastatic potential. This review focuses on updated research about the clinical application of traditional Chinese medicine (TCM) as an adjuvant therapy to lung cancer treatment and the mechanisms of TCM effect on lung cancer in vitro and in vivo.
In this thesis we utilize gene expression data from The Cancer Genome Atlas to identify prognostic biomarkers and predictors of potential response to immunotherapy in lung adenocarcinoma (LUAD) and metastatic skin cutaneous melanoma (SKCM). We utilize various statistical and machine learning techniques in the identification of such biomarker ...
The sensitive A/J strain has a 100% prevalence of spontaneous lung tumors by 18 ±. 24 months of age, with the earliest onset of tumors at. 3 ± 4 months (Shimkin, 1955). The propensity of these ...
1. Introduction. Lung cancer has been the leading cause of cancer death for decades [].From 2007 to 2017, its incidence has increased by 37% [], and the number of deaths attributable to lung cancer was over 130 thousand in 2022 only in the United States (US) [1,2].Due to the asymptomatic nature of early-stage lung cancer, most new cases are diagnosed with advance-stage disease, which often has ...
Non-small-cell lung cancer (NSCLC) is a disease with high morbidity and mortality rates, accounting for approximately 85% of lung cancer cases 1,2.Although Surgery plays a pivotal role in treating ...
The primary focus of this thesis is on cancer cells of different invasive potential, and char-acterization of inherent properties that differ between non-invasive and invasive strains of similar origin. To understand our motivation we will outline some of the main aspects of cancer cells, especially in respect to their survival and spreading.
1 Introduction. Lung cancer is one of the most frequently occurring malignant tumors worldwide and is characterized by a substantially high malignancy and poor prognosis ().According to the latest global cancer statistics, lung cancer remains the leading cause of cancer-related deaths worldwide ().Lung cancer can be histologically classified into two main subtypes: small-cell lung carcinoma ...
In this systematic review and meta-analysis, we evaluated whether lung cancer increases the risk of severe COVID-19 and the risk of dying from the disease. We found that mortality in patients with lung cancer was significantly higher than that in control patients (HR = 2.00 [95%CI 1.52, 2.63], p < 0.01) or with other malignancies (HR = 1.91 [95 ...
The Lung Cancer Master Protocol, or Lung-MAP, is a precision medicine research study for people with advanced non-small cell lung cancer that has continued to grow after treatment. Patients are assigned to different study drug combinations based on the results of genomic profiling of their tumors.
ABOVE: Researchers developed experimental models that mimic the histological transformation from lung adenocarcinoma to small cell lung cancer in mice. Eric Gardner, Weill Cornell Medicine. L ung adenocarcinoma (LUAD), the most common form of lung cancer, is already a challenging disease. It is currently the leading cause of cancer death in the United States. 1 In response to therapies that ...
Background Lung cancer is a malignant tumour with the fastest increase in morbidity and mortality around the world. The clinical treatments available have significant side effects, thus it is desirable to identify alternative modalities to treat lung cancer. Shashen Maidong decoction (SMD) is a commonly used traditional Chinese medicine (TCM) formula for treating lung cancer in the clinic ...
Lung cancer is one of the deadliest types of the disease, claiming the lives of approximately one million people each year. Given the current state of affairs in medicine, it is critical that lung nodule identification be performed on chest CT scans. As a result, the use of CAD systems is crucial for the early detection of lung cancer. ...
Jessica will present the thesis titled "Foundation Model Based Prediction of Lung Cancer Survival Using Temporal Changes in Dual Time Point CT Scans". The thesis committee is chaired by Raymond Mak and includes Benjamin Kann and John Quackenbush. Add to calendar ... PhD '24, studies how children fare when they're born to incarcerated ...
A PhD candidate in the biochemistry and computer science departments, Luopin Wang has been a graduate research assistant in Majid Kazemian's lab since 2018. Now in the final year of her doctoral program, Wang passed her dissertation defense and will be graduating in May with a PhD in computer science. Outstanding Postdoctoral Fellow Award
Non-small cell lung cancer (NSCLC) is one of the most serious diseases affecting human health today, and current research is focusing on gut flora. There is a correlation between intestinal flora imbalance and lung cancer, but the specific mechanism is not clear. Based on the "lung and large intestine being interior-exteriorly related" and ...
Rosemary Rochford, PhD, Received the Henle Award for Decades of Research on the Epstein-Barr Virus-Related Cancer Burkitt lymphoma in sub-Saharan Africa. ... A Combination of Targeted Therapies Proves Effective Against Mutation-Driven Lung Cancer. Mark Harden | April 16, 2024. Cancer Center (SOM) CU Anschutz Anschutz Cancer Pavilion. 1665 North ...