Identifying Primary and Secondary Research Articles
- Primary and Secondary


Primary Research Articles
Primary research articles report on a single study. In the health sciences, primary research articles generally describe the following aspects of the study:
- The study's hypothesis or research question
- Some articles will include information on how participants were recruited or identified, as well as additional information about participants' sex, age, or race/ethnicity
- A "methods" or "methodology" section that describes how the study was performed and what the researchers did
- Results and conclusion section
Secondary Research Articles
Review articles are the most common type of secondary research article in the health sciences. A review article is a summary of previously published research on a topic. Authors who are writing a review article will search databases for previously completed research and summarize or synthesize those articles, as opposed to recruiting participants and performing a new research study.
Specific types of review articles include:
- Systematic Reviews
- Meta-Analysis
- Narrative Reviews
- Integrative Reviews
- Literature Reviews
Review articles often report on the following:
- The hypothesis, research question, or review topic
- Databases searched-- authors should clearly describe where and how they searched for the research included in their reviews
- Systematic Reviews and Meta-Analysis should provide detailed information on the databases searched and the search strategy the authors used.Selection criteria-- the researchers should describe how they decided which articles to include
- A critical appraisal or evaluation of the quality of the articles included (most frequently included in systematic reviews and meta-analysis)
- Discussion, results, and conclusions
Determining Primary versus Secondary Using the Database Abstract
Information found in PubMed, CINAHL, Scopus, and other databases can help you determine whether the article you're looking at is primary or secondary.
Primary research article abstract
- Note that in the "Objectives" field, the authors describe their single, individual study.
- In the materials and methods section, they describe the number of patients included in the study and how those patients were divided into groups.
- These are all clues that help us determine this abstract is describing is a single, primary research article, as opposed to a literature review.
- Primary Article Abstract

Secondary research/review article abstract
- Note that the words "systematic review" and "meta-analysis" appear in the title of the article
- The objectives field also includes the term "meta-analysis" (a common type of literature review in the health sciences)
- The "Data Source" section includes a list of databases searched
- The "Study Selection" section describes the selection criteria
- These are all clues that help us determine that this abstract is describing a review article, as opposed to a single, primary research article.
- Secondary Research Article

- Primary vs. Secondary Worksheet
Full Text Challenge
Can you determine if the following articles are primary or secondary?
- Last Updated: Feb 17, 2024 5:25 PM
- URL: https://library.usfca.edu/primary-secondary
2130 Fulton Street San Francisco, CA 94117-1080 415-422-5555
- Facebook (link is external)
- Instagram (link is external)
- Twitter (link is external)
- YouTube (link is external)
- Consumer Information
- Privacy Statement
- Web Accessibility
Copyright © 2022 University of San Francisco
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
PubMed Central (PMC) Home Page
PubMed Central ® (PMC) is a free full-text archive of biomedical and life sciences journal literature at the U.S. National Institutes of Health's National Library of Medicine (NIH/NLM)
Discover a digital archive of scholarly articles, spanning centuries of scientific research.
Learn how to find and read articles of interest to you.
Collections
Browse the PMC Journal List or learn about some of PMC's unique collections.
For Authors
Navigate the PMC submission methods to comply with a funder mandate, expand access, and ensure preservation.
For Publishers
Learn about deposit options for journals and publishers and the PMC selection process.
For Developers
Find tools for bulk download, text mining, and other machine analysis.
9.8 MILLION articles are archived in PMC.
Content provided in part by:, full participation journals.
Journals deposit the complete contents of each issue or volume.
NIH Portfolio Journals
Journals deposit all NIH-funded articles as defined by the NIH Public Access Policy.
Selective Deposit Programs
Publisher deposits a subset of articles from a collection of journals.
March 21, 2024
Preview upcoming improvements to pmc.
We are pleased to announce the availability of a preview of improvements planned for the PMC website. These…
Dec. 15, 2023
Update on pubreader format.
The PubReader format was added to PMC in 2012 to make it easier to read full text articles on tablet, mobile, and oth…

We are pleased to announce the availability of a preview of improvements planned for the PMC website. These improvements will become the default in October 2024.
NUR 3165 - Nursing Research
- Nursing Databases
- Research Article Basics
- - Practice 1
Finding Primary Research Articles - Overview
- - Practice 2
- Quantitative/Qualitative/Mixed Methods
- - Practice 3
- How to Find Full Text Articles
- Terminology
There are several ways to locate primary research articles as you will see in the following practice exercises (see next page). Here are some tips to consider while looking for original research studies:
Tip #1 - Incorporate subject headings into your search
Subject headings are terms that are part of a controlled vocabulary used to describe the contents tagged inside the article record. These terms can be found in each of the CINAHL Detailed Records under Major Subjects and Minor Subjects. So, if you see the ultimate article, look to see what terms it is tagged with and add them to the search in the appropriate line if relevant. For example,(MH "Emergency Service") is the medical subject heading used for Emergency Department!
To search for possible subject headings, try putting a keyword in a new search and check the Suggest Subject Terms box. The asterisk covers any number of characters (i.e., nurs* yields nurse, nurses, and nurses at the same time). Quotation marks around two or more terms searches them as a phrase.

Try it out! Place the term Hospital Acquired Infection in CINAHL, check the Suggest Subject Terms box and click search to see the subject heading for this term!

Tip #2 - Check the research article box
Databases like CINAHL allow you to select Research Article to retrieve research articles in your search.
Tip #3 - Sections of the Research Article to look for
When reading an article, make sure to look inside the abstract (and the full text) and scan for sections contained in many primary research studies such as Introduction, Participants, Methods, Results and Discussion! Look at those sections to see if the researchers are working directly with the participants and conducting original research.
See the next section for additional tips!
- << Previous: - Practice 1
- Next: - Practice 2 >>
- Last Updated: Mar 11, 2024 4:30 PM
- URL: https://guides.ucf.edu/NUR3165a

Finding Primary Research Articles in the Sciences: Home
- Advanced Search-Databases
- Primary vs. Secondary
- Analyzing a Primary Research Article
- MLA, APA, and Chicago Style
This guide goes over how to find and analyze primary research articles in the sciences (e.g. nutrition, health sciences and nursing, biology, chemistry, physics, sociology, psychology). In addition, the guide explains how to tell the difference between a primary source and a secondary source in scientific subject areas.
If you are looking for how to find primary sources in the humanities and social sciences, such as direct experience accounts in newspapers, diaries, artwork and so forth, please see Finding Primary Sources in the Humanities and Social Sciences .
Recommended Databases
To get started, choose one of the databases below. Once you log in, enter your search terms to start looking for primary articles.
- Link to all Polk State College Library databases
Login Required
You must log in to use library databases and eBooks. When prompted to log in, enter your Passport credentials.
If you have trouble, try resetting your Passport pin , sending an email to [email protected] , or calling the Help Desk at 863.292.3652 .
You can also get help from Ask a Librarian .
Search Tips
Keep your search terms simple.
- No need to type full sentences into the database search box. Limit your search to 2-3 words.
- There is no need to type "research article" into the search box.
Use the "Advanced Search" feature of the database.
- This will allow you to limit your search to only peer reviewed articles or a certain time frame (for example: 2013 or later).
- Click the red tab above for tips on advanced search strategies .
Re-read the assignment guidelines often
- Does this article satisfy the scope of the assignment (e.g. a study focused on nutrition)?
- Does it meet the criteria for the assignment (e.g. an original research article)?
Not finding what you are looking for?
- Ask a Librarian!
Search and Find a Primary Research Article
Are you looking for a primary research journal article if so, that is an article that reports on the results of an original research study conducted by the authors themselves. .
You can use the library's databases to search for primary research articles. A research article will almost always be published in a peer-reviewed journal. Therefore, it is a good idea to limit your results to peer-reviewed articles. Click on the Advanced Search-Databases tab at the top of this guide for instructions.
The following is _not_ primary research:
Review articles are studies that arrive at conclusions after looking over other studies. Therefore, review articles are not primary (think "first") research. There are a variety of review articles, including:
- Literature Reviews
- Systematic Reviews
- Meta-Analyses
- Scoping Reviews
- Topical Reviews
- A review/assessment of the evidence
Having trouble? Look for a method section within the article. If the method section includes the process used to conduct the research, how the data was gathered and analyzed and any limitations or ethical concerns to the study, then it is most likely a primary research article. For example: a research article will describe the number of people (e.g. 175 adults with celiac disease) who participated in the study and who were used to collect data.
If the method section describes how the authors found articles on a topic using search terms or databases , then it is mostly likely a secondary review article and not primary research. If there is no method section, it is not a primary research article.
Other sections in a journal:
Your search may yield these items, too. You can skip these because they are not full write-ups of research:
- Conference Proceedings
- Symposium Publications
Example of a primary research article found in the Library's Academic Search Complete database : (these authors conducted an original research study)
- Lumia et al. (2015) Lumia, M., Takkinen, H., Luukkainen, P., Kaila, M., Lehtinen, J. S., Nwaru, B. I., Tuokkola, J., Niemelä, O., Haapala, A., Ilonen, J., Simell, O., Knip, M., Veijola, R., & Virtanen, S. M. (2015). Food consumption and risk of childhood asthma. Pediatric Allergy & Immunology, 26(8), 789–796. https://doi.org/10.1111/pai.12352
Example of a secondary article found in the Library's Academic Search Complete database : (these authors are reviewing the work of other authors)
- Rachmah et al. (2022) Rachmah, Q., Martiana, T., Mulyono, Paskarini, I., Dwiyanti, E., Widajati, N., Ernawati, M., Ardyanto, Y. D., Tualeka, A. R., Haqi, D. N., Arini, S. Y., & Alayyannur, P. A. (2022). The effectiveness of nutrition and health intervention in workplace setting: A systematic review. Journal of Public Health Research, 11(1), 1–8. https://doi.org/10.4081/jphr.2021.2312
How do I know if this article is primary?
You've found an article in the library databases but how do you know if it's primary .
Look for these sections: (terminology may vary)
- abstract - summarizes paper in one paragraph, states the purpose of the study
- methods - explaining how the experiment was conducted (note: if the method section discusses how a search was conducted that is _not_ primary research)
- results - detailing what happened and providing raw data sets (often as tables or graphs)
- conclusions - connecting the results with theories and other research
- references - to previous research or theories that influenced the research
Scan the article you found to see if it includes the sections above. You don't have to read the full article (yet). Look for the clues highlighted in the images below.

Questions? Use Ask a Librarian
- Next: Advanced Search-Databases >>
- Last Updated: Feb 19, 2024 11:55 AM
- URL: https://libguides.polk.edu/primaryresearch
Polk State College is committed to equal access/equal opportunity in its programs, activities, and employment. For additional information, visit polk.edu/compliance .
- Student Services
- Faculty Services
Peer Review and Primary Literature: An Introduction: Is it Primary Research? How Do I Know?
- Scholarly Journal vs. Magazine
- Peer Review: What is it?
- Finding Peer-Reviewed Articles
- Primary Journal Literature
- Is it Primary Research? How Do I Know?
Components of a Primary Research Study
As indicated on a previous page, Peer-Reviewed Journals also include non -primary content. Simply limiting your search results in a database to "peer-reviewed" will not retrieve a list of only primary research studies.
Learn to recognize the parts of a primary research study. Terminology will vary slightly from discipline to discipline and from journal to journal. However, there are common components to most research studies.
When you run a search, find a promising article in your results list and then look at the record for that item (usually by clicking on the title). The full database record for an item usually includes an abstract or summary--sometimes prepared by the journal or database, but often written by the author(s) themselves. This will usually give a clear indication of whether the article is a primary study. For example, here is a full database record from a search for family violence and support in SocINDEX with Full Text :
Although the abstract often tells the story, you will need to read the article to know for sure. Besides scanning the Abstract or Summary, look for the following components: (I am only capturing small article segments for illustration.)
Look for the words METHOD or METHODOLOGY . The authors should explain how they conducted their research.
NOTE: Different Journals and Disciplines will use different terms to mean similar things. If instead of " Method " or " Methodology " you see a heading that says " Research Design " or " Data Collection ," you have a similar indicator that the scholar-authors have done original research.
Look for the section called RESULTS . This details what the author(s) found out after conducting their research.
Charts , Tables , Graphs , Maps and other displays help to summarize and present the findings of the research.
A Discussion indicates the significance of findings, acknowledges limitations of the research study, and suggests further research.
References , a Bibliography or List of Works Cited indicates a literature review and shows other studies and works that were consulted. USE THIS PART OF THE STUDY! If you find one or two good recent studies, you can identify some important earlier studies simply by going through the bibliographies of those articles.
A FINAL NOTE: If you are ever unclear about whether a particular article is appropriate to use in your paper, it is best to show that article to your professor and discuss it with them. The professor is the final judge since they will be assigning your grade.
Subject Guide

- << Previous: Primary Journal Literature
- Last Updated: Nov 16, 2022 12:46 PM
- URL: https://suffolk.libguides.com/PeerandPrimary

Animal Science
How to identify peer reviewed journals, how to identify primary research articles.
- Reference Sources
- Key Journals
- Writing & Citing
- Self Checkout
- Anatomy Study Resources
- Peer Reviewed Journals Quiz How do I know if a journal is peer reviewed? What is peer review, anyway? Take this short quiz to test your knowledge and perhaps learn something new!
- Primary Research Articles Quiz How do I know if an article is a primary or secondary research article? Are there search techniques that will help me find them? Take this short quiz to test your knowledge and perhaps learn something new!
You must get all answers correct to submit the quiz!
Peer review is defined as “a process of subjecting an author’s scholarly work, research or ideas to the scrutiny of others who are experts in the same field” ( 1 ). Peer review is intended to serve two purposes:
- It acts as a filter to ensure that only high quality research is published, especially in reputable journals, by determining the validity, significance and originality of the study.
- Peer review is intended to improve the quality of manuscripts that are deemed suitable for publication. Peer reviewers provide suggestions to authors on how to improve the quality of their manuscripts, and also identify any errors that need correcting before publication.
How do you determine whether an article qualifies as being a peer-reviewed journal article?
- If you're searching for articles in certain databases, you can limit your search to peer-reviewed sources simply by selecting a tab or checking a box on the search screen.
- If you have an article, an indication that it has been through the peer review process will be the publication history , usually at the beginning or end of the article.
- If you're looking at the journal itself, go to the editorial statement or instructions to authors (usually in the first few pages of the journal or at the end) for references to the peer-review process.
- Lookup the journal by title or ISSN in the ProQuest Source Evaluation Aid .
- Careful! Not all information in a peer-reviewed journal is actually reviewed. Editorials, letters to the editor, book reviews, and other types of information don't count as articles, and may not be accepted by your professor.
What about preprint sites and ResearchGate?
- A preprint is a piece of research that has not yet been peer reviewed and published in a journal. In most cases, they can be considered final drafts or working papers. Preprint sites are great sources of current research - and most preprint sites will provide a link to a later, peer-reviewed version of an article.
- ResearchGate is a commercial social networking site for scientists and researchers to share papers, ask and answer questions, and find collaborators. Members can upload research output including papers, chapters, negative results, patents, research proposals, methods, presentations, etc. Researchers can access these materials, and also contact members to ask for access to material that has not been shared, usually because of copyright restrictions. There is a filter to limit results to articles, but it can be difficult to determine the publication history of ResearchGate items and whether they have been published in peer reviewed sources.
A primary research article reports on an empirical research study conducted by the authors. The goal of a primary research article is to present the result of original research that makes a new contribution to the body of knowledge.
Characteristics:
- Almost always published in a peer-reviewed journal
- Asks a research question or states a hypothesis or hypotheses
- Identifies a research population
- Describes a specific research method
- Tests or measures something
- Often (but not always) structured in a standard format called IMRAD: Introduction, Methods, Results, and Discussion
- Words to look for as clues include: analysis, study, investigation, examination, experiment, numbers of people or objects analyzed, content analysis, or surveys.
To contrast, the following are not primary research articles (i.e., they are secondary sources):
- Literature reviews/Review articles
- Meta-Analyses (studies that arrive at conclusions based on research from many other studies)
- Editorials & Letters
- Dissertations
Articles that are NOT primary research articles may discuss the same research, but they are not reporting on original research, they are summarizing and commenting on research conducted and published by someone else. For example, a literature review provides commentary and analysis of research done by other people, but it does not report the results of the author's own study and is not primary research.
- << Previous: Home
- Next: Reference Sources >>
- Last Updated: Aug 24, 2023 2:38 PM
- URL: https://libguides.berry.edu/ans
How to Find Primary Research Articles on Google Scholar

How to Find Primary Research Articles on Google Scholar can be a daunting task. But with the right tips and tricks, you can quickly locate relevant sources to inform your work or study. By leveraging advanced search features like My Library, you’ll be able to stay organized while exploring topics of interest in no time. Let’s dive into how best to find primary research articles on Google Scholar so that you can get started uncovering valuable insights today.
Table of Contents
What is Google Scholar?
Searching for primary research articles on google scholar, tips for effective searches on google scholar, utilizing advanced search features, keeping track of your research with my library on google scholar, additional resources for finding primary research articles on google scholar, faqs in relation to how to find primary research articles on google scholar, how do i search for only primary articles in google scholar, how do i find primary research articles, how do i find research articles on google scholar, how do you tell if an article is a primary or secondary source.
Google Scholar is an online search engine that allows users to find primary research articles. Google Scholar, established in 2004, is a powerful search engine that gives access to scholarly documents including theses, preprints, and books. By using advanced algorithms and natural language processing techniques it offers a more comprehensive view of academic publications than traditional databases or search engines like Google.
How to Find Primary Research Articles on Google Scholar has numerous advantages; it provides a convenient way for researchers to quickly find applicable sources needed for their research without having to browse through many web pages or databases. Secondly, its sophisticated algorithms allow researchers to refine their searches based on relevance and date published to easily narrow down results for specific topics or time periods. Finally, because it indexes content from across the web – including open-access repositories such as PubMed Central – users have access to full-text versions of articles that may not be available elsewhere.
Accessing Google Scholar is easy; simply go to scholar.google.com and start searching with keywords related to your topic area or use the Advanced Search feature if you want more control over your results (e.g., restricting by author name). You can also sign up for an account which will enable you to save searches, create alerts when new content is added that matches your criteria, and organize references into collections known as ‘My Library’ – making tracking progress on a project much more efficient.
Google Scholar is an invaluable resource for researchers looking to access primary research articles. With the right search techniques, you can easily find full-text articles on Google Scholar and maximize your research potential. Next, we’ll explore how to use the search interface and refine results in order to locate these resources more effectively.
“Easily find primary research articles for your #R&D project with Google Scholar. Advanced algorithms and natural language processing make it easier to narrow down results quickly.” #Cypris Click to Tweet
To make the process easier, it is important to understand the search interface and refine your results with filters and preferences.
The first step in searching for primary research articles on Google Scholar is understanding the search interface. This includes learning how to use keywords, Boolean operators (AND, OR, NOT), quotation marks (” “) for exact phrases, and wildcards (*). These search parameters can be employed to refine the results, making them pertinent to your inquiry.
Utilizing filters and personal preferences to narrow down search results can expedite the discovery of what is needed. With advanced features like citation tracking, “My Library” which allows users to save their searches, and “Similar Articles” for discovering related topics within a field of study, the research process is made easier. Additionally, keywords such as Boolean operators (AND, OR NOT), quotation marks (” “) for exact phrases, and wildcards (*) can be used to narrow down results in order to make them more relevant.
Finally, finding full-text articles is key when researching primary research papers on Google Scholar. The platform offers access to free versions of some publications through its “Find Full Text @ Your Library” feature but many require a subscription or purchase fee before viewing them in full detail online or downloading them as PDFs.
Exploring Google Scholar for primary research articles can be laborious, yet with some useful tips and tricks you can enhance your search results. Now that we have an understanding of the search interface, let’s explore how to refine our results and find full-text articles using advanced features such as filters and preferences.
Unlock the power of Google Scholar for primary research papers with advanced features like citation tracking, My Library, and Similar Articles. Use Boolean operators & wildcards to refine your search results. #GoogleScholar #ResearchPapers Click to Tweet
Google Scholar is an invaluable tool for researchers, scientists, and engineers looking to stay up-to-date on the latest research in their field. With its advanced search features, it can help you quickly find primary research articles that are relevant to your project or interests. Here are some suggestions to optimize your utilization of Google Scholar when seeking out primary research papers.
Google Scholar has several advanced search options that allow you to refine your searches and find more specific results. For example, you can limit your search by date range, language, author name, or journal title. Boolean operators, like “AND” and “OR”, can be utilized to form a single query by combining various keywords.

To refine your search even further, you can utilize the filters and preferences available on Google Scholar to narrow down results according to peer-reviewed papers from journals with high-impact factors or exclude certain authors or topics. For instance, if you want only peer-reviewed papers from journals with high-impact factors then simply select those filters before conducting your search. Additionally, if there are certain authors or topics that you would like excluded from your results then this too can be done via the preferences menu within Google Scholar.
Once you have located some applicable articles through basic keyword searches, delving into associated citations and related content can help to expand your understanding of the topic. This is especially helpful if there is not much information available on a particular subject yet, but still offers potential avenues of exploration worth pursuing further down the line. By exploring related articles and citations associated with each article one will often uncover new ideas which could potentially lead them toward interesting discoveries.
By making use of the sophisticated search capabilities, filters, and preferences provided by Google Scholar, one can easily identify primary research material related to their requirements. My Library on Google Scholar is an excellent tool for organizing and tracking your research; let’s explore how it works.
Key Takeaway Google Scholar provides advanced search features, filters and preferences to help researchers quickly locate primary research articles relevant to their project or interests. By making use of these tools and exploring related articlescitations associated with each article one can uncover new ideas that could lead them towards interesting discoveries. Google Scholar is a great aid in locating pertinent research articles.
My Library on Google Scholar is a great asset for scientists and innovators to monitor their research progress. My Library enables users to construct a personalized repository of scholarly works, which they can organize into categories, export as bibliographies, or share with others.
Setting up a personal library in My Library is easy. To create a personal library, simply click the “My Library” link at the top right corner of any page on Google Scholar and select “Create new library” from the drop-down menu. Once your library has been created, you can start adding articles by clicking the “Save” button next to each article title in your search results list.
Organizing your library is also simple; simply drag and drop articles into different folders within My Library for easy access later on. You can also create collections of related topics or research themes which are great for organizing large amounts of data quickly and easily. Moreover, you can label articles with descriptors to make them easier to locate when needed.
By utilizing My Library on Google Scholar, researchers can easily keep track of their research and stay organized. Additionally, by exploring other databases in conjunction with Google Scholar as well as open-access journals and interlibrary loan services, they can find even more primary research articles to further their studies.
Key Takeaway My Library on Google Scholar is a great resource for researchers and innovators to stay organized with their research. Creating a library is straightforward – just hit the ‘Create new library’ button in the top right of any page on Google Scholar, and then drag & drop articles into collections or folders to keep them ordered. Moreover, you can assign labels or tags to make it simpler to locate the material when necessary.
It can provide access to a wide variety of sources, including journal articles, books, and conference papers. Nevertheless, in order to broaden one’s search range, other databases and sources can be used alongside Google Scholar.
Using Other Databases in Conjunction with Google Scholar: Many academic institutions have their own subscription-based library databases that can be accessed through the institution’s website or portal. These databases may include full-text versions of some journals not available on Google Scholar as well as more comprehensive indexing than what is available on Google Scholar. Moreover, numerous universities offer access to specialized databases such as Web of Science or Scopus that enable users to search across multiple areas and sources simultaneously.
Open-access journals, which receive funding from sources such as the NIH and Wellcome Trusts, provide free online content under Creative Commons licenses for readers to share or reuse without permission. Open-access journals typically make all content freely available online and often use Creative Commons licenses so readers are free to share and reuse material without permission from the publisher or author(s). While these publications tend to focus more heavily on scientific topics rather than humanities topics they still contain valuable information worth exploring when searching for primary research articles related specifically to science fields such as biology or medicine.
If a desired article cannot be located elsewhere, interlibrary loan services may provide an avenue to acquire it through either physical or digital means. Through this service, users can request copies of materials held by another library either physically (through mail) or electronically (via email). This allows researchers who do not have immediate access to certain materials due to geographical restrictions the ability to acquire them nonetheless, thus greatly expanding their research capabilities beyond what would otherwise be possible with just local resources alone.
Key Takeaway Google Scholar is a great tool for finding primary research articles, however there are other databases and resources that can be used in conjunction with it to maximize search capabilities. Additionally, open access journals may provide valuable content related to scientific fields while interlibrary loan services can also help researchers acquire materials from libraries located elsewhere.
To search for primary articles in Google Scholar, first, go to the main page and select ‘Advanced Search’. In the Advanced Search window, check off the box that says ‘Only show results from content I can access’ and then select ‘Include Patents’. Finally, click on ‘Search’. This will filter out all secondary sources such as reviews or books, leaving only primary research articles relevant to your query.
Primary research materials can be obtained through multiple avenues, such as searching online repositories, utilizing sophisticated search strategies, and consulting specialists in the discipline. Utilizing PubMed and other online databases, researchers can access an abundance of primary research articles covering a broad range of topics. Advanced search techniques involve combining keywords with Boolean operators (AND/OR) to refine searches for specific results. Consulting experts in the field is also an effective way to locate relevant primary research articles as they have specialized knowledge about certain areas that may not be available from other sources.
Begin your hunt for research articles on Google Scholar by inputting a keyword or phrase in the search field. You can refine your search results by applying filters such as date of publication, author name, and topic area. To further narrow down your search results you can use advanced search features like exact phrases and multiple keywords. Additionally, you may access scholarly literature through library databases that are connected to Google Scholar. Finally, save time by setting up email alerts for newly published papers related to topics of interest.
A primary source is an original document or record that provides first-hand information about a particular topic. Examples of primary sources can include interviews, diaries, letters, articles from when an event occurred, and photos and videos taken during the occurrence. Secondary sources are documents or records created after the fact by someone who did not experience the events firsthand. These may include books, journal articles, and reviews that analyze or discuss research already published by others.
How to find primary research articles on Google Scholar is an essential skill for researchers and innovators. With its advanced search capabilities, My Library feature, and additional resources available online, it can be an invaluable asset in the quest to discover new insights into any given topic. Whether you are looking for one article or hundreds of them on a specific subject matter – Google Scholar is here to help. Use these tips as your guide when searching for primary research articles on Google Scholar so that you can get the most out of this platform’s features.
Discover the power of Cypris to quickly find primary research articles on Google Scholar and unlock insights faster for your R&D and innovation teams. Unlock time-saving solutions with our comprehensive platform that centralizes data sources into one easy-to-use interface.
Similar insights you might enjoy

Gallium Nitride Innovation Pulse

Carbon Capture & Storage Innovation Pulse

Sodium-Ion Batteries Innovation Pulse
PubMed: Find Research Articles
- Run a Search
- Find Full Text
- Refine Searches
- Find Research Articles
- MeSH/Advanced Search
- Export to Citation Managers
- New PubMed Essentials
Finding Comparative Effectiveness Research
Comparative effectiveness research is the conduct and synthesis of research comparing the benefits and harms of different interventions and strategies to prevent, diagnose, treat and monitor health conditions in "real world" settings.
Two specialized resources are available to inform comparative effectiveness research:
Comparative Effectiveness Research on the PubMed Topic-Specific Queries page. Provides specialized PubMed searches of published research and research in progress to help inform investigations of comparative effectiveness.
- Medline Plus is the world’s largest medical library, it brings you information about diseases, conditions, and wellness issues in language you can understand. MedlinePlus offers reliable, up-to-date health information, anytime, anywhere, for free.
3 Ways to Find Research Articles in PubMed
1. filter (limit) to article type.
Most citations in PubMed are for journal articles. However, you may limit your retrieval based on the type of material the article represents. Use the Filters on the Results page sidebar and look at the Article Types checklist which contains a list of frequently searched publication types.
For example, choose Randomized Controlled Trial or Clinical Trial or Meta-Analysis from the list.
2. PubMed Clinical Queries
Enter your search terms and evidence-filtered citations will appear under Clinical Study Categories. Systematic Reviews or Medical Genetics. The Clinical Queries link is found on the PubMed home page or under the More Resources drop-down at the top of the Advanced Search page.
The resulting retrieval in PubMed Clinical Queries can be further refined using PubMed's Filters, e.g., English language, humans.
3. Limit to Articles with Structured Abstracts
Many abstracts that are added to PubMed include section labels such as BACKGROUND, OBJECTIVE, METHODS, RESULTS, and CONCLUSIONS. These 'structured' abstracts appear in many different article types such as review articles, original research, and practice guidelines and facilitate skimming of citations for relevance and specific information such as research design within the Methods section. The presence of structured abstracts in citations are a searchable feature in PubMed. To limit to citations containing structured abstracts, include the term hasstructuredabstract in the search box.
For example: valerian AND sleep AND hasstructuredabstract
- << Previous: Refine Searches
- Next: MeSH/Advanced Search >>
- Last Updated: Jul 18, 2023 11:35 AM
- URL: https://guides.lib.vt.edu/pubmed_tips
- Harvard Library
- Research Guides
- Faculty of Arts & Sciences Libraries
- Identifying Articles
- PubMed at Harvard
- Searching in PubMed
- My NCBI in PubMed
- Utilizing Search Results
- Scenarios in PubMed
Primary Research Article
Review article.
Identifying and creating an APA style citation for your bibliography:
- Author initials are separated by a period
- Multiple authors are separated by commas and an ampersand (&)
- Title format rules change depending on what is referenced
- Double check them for accuracy

Identifying and creating an APA style in-text citation:
- eg. (Smith, 2022) or (Smith & Stevens, 2022)
The structure of this changes depending on whether a direct quote or parenthetical used:
Direct Quote: the citation must follow the quote directly and contain a page number after the date
eg. (Smith, 2022, p.21)
Parenthetical: the page number is not needed
For more information, take a look at Harvard Library's Citation Styles guide !
A primary research article typically contains the following section headings:
"Methods"/"Materials and Methods"/"Experimental Methods"(different journals title this section in different ways)
"Results"
"Discussion"
If you skim the article, you should find additional evidence that an experiment was conducted by the authors themselves.
Primary research articles provide a background on their subject by summarizing previously conducted research, this typically occurs only in the Introduction section of the article.
Review articles do not report new experiments. Rather, they attempt to provide a thorough review of a specific subject by assessing either all or the best available scholarly literature on that topic.
Ways to identify a review article:
- Author(s) summarize and analyze previously published research
- May focus on a specific research question, comparing and contrasting previously published research
- Overview all of the research on a particular topic
- Does not contain "methods" or "results" type sections
- << Previous: Scenarios in PubMed
- Last Updated: Oct 3, 2023 4:16 PM
- URL: https://guides.library.harvard.edu/PubMed
Harvard University Digital Accessibility Policy
Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, generate accurate citations for free.
- Knowledge Base
- Working with sources
How to Find Sources | Scholarly Articles, Books, Etc.
Published on June 13, 2022 by Eoghan Ryan . Revised on May 31, 2023.
It’s important to know how to find relevant sources when writing a research paper , literature review , or systematic review .
The types of sources you need will depend on the stage you are at in the research process , but all sources that you use should be credible , up to date, and relevant to your research topic.
There are three main places to look for sources to use in your research:
Research databases
- Your institution’s library
- Other online resources
Table of contents
Library resources, other online sources, other interesting articles, frequently asked questions about finding sources.
You can search for scholarly sources online using databases and search engines like Google Scholar . These provide a range of search functions that can help you to find the most relevant sources.
If you are searching for a specific article or book, include the title or the author’s name. Alternatively, if you’re just looking for sources related to your research problem , you can search using keywords. In this case, it’s important to have a clear understanding of the scope of your project and of the most relevant keywords.
Databases can be general (interdisciplinary) or subject-specific.
- You can use subject-specific databases to ensure that the results are relevant to your field.
- When using a general database or search engine, you can still filter results by selecting specific subjects or disciplines.
Example: JSTOR discipline search filter

Check the table below to find a database that’s relevant to your research.
Google Scholar
To get started, you might also try Google Scholar , an academic search engine that can help you find relevant books and articles. Its “Cited by” function lets you see the number of times a source has been cited. This can tell you something about a source’s credibility and importance to the field.
Example: Google Scholar “Cited by” function
Boolean operators
Boolean operators can also help to narrow or expand your search.
Boolean operators are words and symbols like AND , OR , and NOT that you can use to include or exclude keywords to refine your results. For example, a search for “Nietzsche NOT nihilism” will provide results that include the word “Nietzsche” but exclude results that contain the word “nihilism.”
Many databases and search engines have an advanced search function that allows you to refine results in a similar way without typing the Boolean operators manually.
Example: Project Muse advanced search

The only proofreading tool specialized in correcting academic writing - try for free!
The academic proofreading tool has been trained on 1000s of academic texts and by native English editors. Making it the most accurate and reliable proofreading tool for students.
Try for free
You can find helpful print sources in your institution’s library. These include:
- Journal articles
- Encyclopedias
- Newspapers and magazines
Make sure that the sources you consult are appropriate to your research.
You can find these sources using your institution’s library database. This will allow you to explore the library’s catalog and to search relevant keywords. You can refine your results using Boolean operators .
Once you have found a relevant print source in the library:
- Consider what books are beside it. This can be a great way to find related sources, especially when you’ve found a secondary or tertiary source instead of a primary source .
- Consult the index and bibliography to find the bibliographic information of other relevant sources.
You can consult popular online sources to learn more about your topic. These include:
- Crowdsourced encyclopedias like Wikipedia
You can find these sources using search engines. To refine your search, use Boolean operators in combination with relevant keywords.
However, exercise caution when using online sources. Consider what kinds of sources are appropriate for your research and make sure the sites are credible .
Look for sites with trusted domain extensions:
- URLs that end with .edu are educational resources.
- URLs that end with .gov are government-related resources.
- DOIs often indicate that an article is published in a peer-reviewed , scientific article.
Other sites can still be used, but you should evaluate them carefully and consider alternatives.
If you want to know more about ChatGPT, AI tools , citation , and plagiarism , make sure to check out some of our other articles with explanations and examples.
- ChatGPT vs human editor
- ChatGPT citations
- Is ChatGPT trustworthy?
- Using ChatGPT for your studies
- What is ChatGPT?
- Chicago style
- Paraphrasing
Plagiarism
- Types of plagiarism
- Self-plagiarism
- Avoiding plagiarism
- Academic integrity
- Consequences of plagiarism
- Common knowledge
Scribbr Citation Checker New
The AI-powered Citation Checker helps you avoid common mistakes such as:
- Missing commas and periods
- Incorrect usage of “et al.”
- Ampersands (&) in narrative citations
- Missing reference entries
You can find sources online using databases and search engines like Google Scholar . Use Boolean operators or advanced search functions to narrow or expand your search.
For print sources, you can use your institution’s library database. This will allow you to explore the library’s catalog and to search relevant keywords.
It is important to find credible sources and use those that you can be sure are sufficiently scholarly .
- Consult your institute’s library to find out what books, journals, research databases, and other types of sources they provide access to.
- Look for books published by respected academic publishing houses and university presses, as these are typically considered trustworthy sources.
- Look for journals that use a peer review process. This means that experts in the field assess the quality and credibility of an article before it is published.
When searching for sources in databases, think of specific keywords that are relevant to your topic , and consider variations on them or synonyms that might be relevant.
Once you have a clear idea of your research parameters and key terms, choose a database that is relevant to your research (e.g., Medline, JSTOR, Project MUSE).
Find out if the database has a “subject search” option. This can help to refine your search. Use Boolean operators to combine your keywords, exclude specific search terms, and search exact phrases to find the most relevant sources.
There are many types of sources commonly used in research. These include:
You’ll likely use a variety of these sources throughout the research process , and the kinds of sources you use will depend on your research topic and goals.
Scholarly sources are written by experts in their field and are typically subjected to peer review . They are intended for a scholarly audience, include a full bibliography, and use scholarly or technical language. For these reasons, they are typically considered credible sources .
Popular sources like magazines and news articles are typically written by journalists. These types of sources usually don’t include a bibliography and are written for a popular, rather than academic, audience. They are not always reliable and may be written from a biased or uninformed perspective, but they can still be cited in some contexts.
Cite this Scribbr article
If you want to cite this source, you can copy and paste the citation or click the “Cite this Scribbr article” button to automatically add the citation to our free Citation Generator.
Ryan, E. (2023, May 31). How to Find Sources | Scholarly Articles, Books, Etc.. Scribbr. Retrieved April 15, 2024, from https://www.scribbr.com/working-with-sources/finding-sources/
Is this article helpful?

Eoghan Ryan
Other students also liked, types of sources explained | examples & tips, primary vs. secondary sources | difference & examples, boolean operators | quick guide, examples & tips, "i thought ai proofreading was useless but..".
I've been using Scribbr for years now and I know it's a service that won't disappoint. It does a good job spotting mistakes”
Font Awesome fontawesome.com --> We are making changes to how you access electronic resources. Learn more.
How do I find Primary Sources as a science or social science student?
Primary Sources are:
- Researchers reporting first-hand about their new research
- Includes some Journal articles and some Books (monographs)
In contrast, Secondary Sources summarize, analyze or report the work of other researchers.
The most common type of journal article you will find in the sciences deals with primary research . These articles describe an original experiment or analysis that adds to current knowledge a particular topic.
Typically, Primary journal articles will have a common structure that includes:
- Introduction
- Methods/Methods & Materials
- Results
- Discussion
- Conclusion.
Look for a Methods or Methods and Materials Section as a quick check to see if an article may be primary. Read this section to see if the researchers are talking about their new research.
Can't find what you're looking for? Contact us.

Understanding Nursing Research
What is primary research, how can i tell if my article is "primary research", limiting your search to primary research.
- Qualitative vs. Quantitative Research
- Experimental Design
- Is it a Nursing journal?
- Is it Written by a Nurse?
- Systematic Reviews and Secondary Research
- Quality Improvement Plans
Your Team! College of Education and Human Development and College of Nursing and Health Sciences

Left to Right: Trisha Hernandez, Emily Murphy, Lorin Flores, Aida Almanza-Ferro.
We are the librarians for College of Education and Human Development, and the College of Nursing. We look forward to working with you! To contact us or to make an appointment:
Submit your request and we'll get right back to you!
Or, you can reach out directly. For our email addresses and phone numbers, see the list below:
Aida Almanza-Ferro | [email protected] | 361-825-2356 Lorin Flores | [email protected] | 361-825-2609 Trisha Hernandez | [email protected] |361-825-2687 Emily Sartorius Murphy | [email protected] | 361-825-2610 Librarians are available M-F, 8-5.
The phrase "Primary" can mean something different depending on what subject you're in.
In History , for example, you might hear the phrase "primary sources." This means the researcher is looking for sources that date back to when an event occurred. Primary sources can be a diary, a photograph, or a newspaper clipping.
If this is the kind of research you're looking for, check out this research guide on how to find primary sources:
- Primary Sources
If you're in Nursing or another scientific field you're more likely to hear the phrase "Primary Research."
Primary Research refers to research that was conducted by the author of the article you're reading. So if you're reading an article and in the methodology section the author refers to recruiting participants, identifying a control group, etc. you can be pretty sure the author has conducted the research themselves.
When you're asked to find primary research, you're being asked to find articles describing research that was conducted by the authors.
Check out the video below for an explanation of the differences between primary and secondary research.
To determine if the article you're looking at is considered Primary Research, look for the following:
- In the Abstract, can you find a description of research being conducted?
- Were participants recruited?
- Were surveys distributed?
The main question to ask yourself is "Did the author conduct research, or did they read and synthesize other people's research?"
If you've found an article in CINAHL and you want to know if it's primary research, look under "Publication Type" to see if it's a research article.

This is not always 100% correct, though. To be sure, you should always read the Methodology section to understand what kind of article you're looking at.
If you're using PubMed, you can check the article's Keywords and Abstract for clues to see if the article is primary research, like in the article below:

Or you can check to see if the article includes a "Publication Type" section like this article:

The following Publication Types are usually considered Primary Research:
- Adaptive Clinical Trial
- Clinical Study
- Clinical Trial
- Controlled Clinical Trial
- Equivalence Trial
- Evaluation Studies
- Observational Study
- Pragmatic Clinical Trial
- Randomized Controlled Trial
Remember, you will always need to read the Methodologies section of an article to be sure the article is an example of primary research!
In certain databases you can specify that you're only interested in resources that are considered primary research.
Two of those databases are CINAHL and PubMed, which you can access here:
To limit your results to primary research in CINAHL, check the "Research Article" box on the homepage before you hit "Search"

This check box is helpful, but it isn't 100% correct, so always read the Methodology section of your article to determine what kind of article it is!
If you're conducting a search in PubMed and want to limit your results to a certain kind of article, you can enter your search terms on the homepage and click "Search."
Then, when you're on your results page, use the limiters on the left side of the screen to specify the "Article Type" you're interested in. Under "Article Types" click the "Customize..." link to see the full list of article types available to you.

Check any of the article types you're interested in (don't forget to scroll down on this list!) and then click the blue "Show" button at the bottom of the pop up window.
Now the Article Types you just selected should appear under the Article Types heading. Click on the article types you want to show up in your results list and your results will limit themselves to just those that meet your criteria.

Remember to read the article's Methodology section yourself before deciding whether or not it's Primary Research! These limits are great, but they aren't always 100% accurate.
- Next: Qualitative vs. Quantitative Research >>
- Last Updated: Feb 6, 2024 9:34 AM
- URL: https://guides.library.tamucc.edu/nursingresearch
Information Technology
Primary research articles.
- Library vs. Google
- Background Reading
- Keyword Searching
- Evaluating Sources
- Citing Sources
- Need more help?
How Can I Find Primary Research Articles?
Many of the recommended databases in this subject guide contain primary research articles (also known as empirical articles or research studies). Search in databases like ScienceDirect , MEDLINE , and Health Source: Nursing/Academic Edition .
Primary Research Articles: How Will I Know One When I See One?
Primary research articles to conduct and publish an experiment or research study, an author or team of authors designs an experiment, gathers data, then analyzes the data and discusses the results of the experiment. a published experiment or research study will therefore look very different from other types of articles (newspaper stories, magazine articles, essays, etc.) found in our library databases. the following guidelines will help you recognize a primary research article, written by the researchers themselves and published in a scholarly journal., structure of a primary research article typically, a primary research article has the following sections:.
- The author summarizes her article
- The author discusses the general background of her research topic; often, she will present a literature review, that is, summarize what other experts have written on this particular research topic
- The author describes the study she designed and conducted
- The author presents the data she gathered during her experiment
- The author offers ideas about the importance and implications of her research findings, and speculates on future directions that similar research might take
- The author gives a References list of sources she used in her paper
The structure of the article will often be clearly shown with headings: Introduction, Method, Results, Discussion.
A primary research article will almost always contains statistics, numerical data presented in tables. Also, primary research articles are written in very formal, very technical language.
- << Previous: Resources
- Next: Research Tips >>
- Last Updated: Jan 4, 2024 11:48 AM
- URL: https://libguides.umgc.edu/information-technology
- UNC Libraries
- Collections
- Creative Music Research in Special Collections
Primary Source Analysis
Creative music research in special collections: primary source analysis.
- Archives and Libraries
- Using a Finding Aid
- Registering & Requesting Materials
Introduction to Primary Sources
- Music Copyright
- Creative Research Opportunities
- Creative Music Research Examples and Methodologies
What Are Primary Sources?
Primary sources are materials that were created during the time in question. They are the evidence of a particular time and place and moment. Secondary sources provide analysis of other materials, but primary sources are the raw and unfiltered data. Unlike secondary sources which already provide an interpretation of something, working with primary sources forces the researcher to conduct their own analysis. Examples of primary sources include letters, dairies, newspapers, original musical scores, audio and video recordings, oral histories, photographs and more.
Primary source analysis asks researches to observe, reflect and question the materials, thinking about criteria such as
- Materiality
- Purpose and Audience
More more information on primary source analysis, visit Library of Congress Primary Source Guides and Analysis Tools .
Practice Primary Source Analysis
Let's use recorded sound as an example to conduct a primary source analysis..
Analyzing a sound recording poses its own questions and challenges. There can be multiple layers of content on a music recording. For instance, you may notice the sounds created by the performers, the sounds created by other people present and perhaps background noise created by the recording technology. There is also the description of the sound recording which may or may not accurately depict what and who is on the recording itself. Additionally, there is the “liveness” of performance to consider – how does environment and context affect a live performance?
Listen: SFC Audio Cassette FS-20009/12936, Elizabeth Cotten Birthday, 6 January 1979; Elizabeth Cotton part 1, 10 January 1979: Side 1
What do you know about the recording before listening to it.
- What is the materiality of the recording itself? What technology was used to record it?
- What is in the description (date, location, personnel, content, etc)
What do you hear in the recording?
- What is the first thing you notice?
- What is the content of the recording? Are there sounds in addition to this content?
- Are there people present in the recording that aren’t listed in the description? If so, who are they?
How does the recording make you feel?
- What emotions are evoked when listening to the recording?
- What role does emotion play in your interpretation of the performance?
- How do you think the performers are feeling in the recording?

What is the context of the recording? When/where was it recorded?
- What is the context for the sound recording? Who recorded it and why? For whom?
- What is the relationship between the persons being recorded and the person doing the recording? Is this relationship described?
- What is the relationship between the musicians and the content being performed?
What does this recording tell you about the artists' creative process?
- What is distinct about this recording compared to other contemporary commercial recordings?
- What artistic processes do you hear in the recording? Are there stops and starts? Is it rehearsed or impromptu?
- How do you think the context of the recording impacts the "liveness" of the musical performance?
- What techniques do you hear that are unique to this performance? Does the performance style differ from techniques you are familiar with?
Continued Learning
- What other information would be helpful in understanding the context?
- Have the musicians been recorded in other contexts?
- What other musicians recorded in the same region around the same time, or in different time periods? Have other musicians recorded the same repertoire?
- What else was happening around the time and place that the sound recording was made?
- Downloadable PDF of Primary Source Analysis for Music Recordings
Primary Source Analysis & Performance
How can primary source analysis enrich creative practice.
Analyzing primary sources can give us insight into the creative process. Unlike published recordings, primary sources can show the process rather than the product. Perhaps there are rehearsal notes, recordings, documentation of conversations around the performance, etc. These can inform our own creative practices.
- << Previous: Registering & Requesting Materials
- Next: Copyright & Permissions >>
- Last Updated: Apr 15, 2024 10:30 AM
- URL: https://guides.lib.unc.edu/musicresearch
Search & Find
- E-Research by Discipline
- More Search & Find
Places & Spaces
- Places to Study
- Book a Study Room
- Printers, Scanners, & Computers
- More Places & Spaces
- Borrowing & Circulation
- Request a Title for Purchase
- Schedule Instruction Session
- More Services
Support & Guides
- Course Reserves
- Research Guides
- Citing & Writing
- More Support & Guides
- Mission Statement
- Diversity Statement
- Staff Directory
- Job Opportunities
- Give to the Libraries
- News & Exhibits
- Reckoning Initiative
- More About Us
- Search This Site
- Privacy Policy
- Accessibility
- Give Us Your Feedback
- 208 Raleigh Street CB #3916
- Chapel Hill, NC 27515-8890
- 919-962-1053
- Open access
- Published: 15 April 2024
Machine-learning analysis reveals an important role for negative selection in shaping cancer aneuploidy landscapes
- Juman Jubran 1 na1 ,
- Rachel Slutsky 2 na1 ,
- Nir Rozenblum 2 ,
- Lior Rokach 3 ,
- Uri Ben-David 2 na2 &
- Esti Yeger-Lotem ORCID: orcid.org/0000-0002-8279-7898 1 , 4 na2
Genome Biology volume 25 , Article number: 95 ( 2024 ) Cite this article
5 Altmetric
Metrics details
Aneuploidy, an abnormal number of chromosomes within a cell, is a hallmark of cancer. Patterns of aneuploidy differ across cancers, yet are similar in cancers affecting closely related tissues. The selection pressures underlying aneuploidy patterns are not fully understood, hindering our understanding of cancer development and progression.
Here, we apply interpretable machine learning methods to study tissue-selective aneuploidy patterns. We define 20 types of features corresponding to genomic attributes of chromosome-arms, normal tissues, primary tumors, and cancer cell lines (CCLs), and use them to model gains and losses of chromosome arms in 24 cancer types. To reveal the factors that shape the tissue-specific cancer aneuploidy landscapes, we interpret the machine learning models by estimating the relative contribution of each feature to the models. While confirming known drivers of positive selection, our quantitative analysis highlights the importance of negative selection for shaping aneuploidy landscapes. This is exemplified by tumor suppressor gene density being a better predictor of gain patterns than oncogene density, and vice versa for loss patterns. We also identify the importance of tissue-selective features and demonstrate them experimentally, revealing KLF5 as an important driver for chr13q gain in colon cancer. Further supporting an important role for negative selection in shaping the aneuploidy landscapes, we find compensation by paralogs to be among the top predictors of chromosome arm loss prevalence and demonstrate this relationship for one paralog interaction. Similar factors shape aneuploidy patterns in human CCLs, demonstrating their relevance for aneuploidy research.
Conclusions
Our quantitative, interpretable machine learning models improve the understanding of the genomic properties that shape cancer aneuploidy landscapes.
Introduction
Aneuploidy, defined as an abnormal number of chromosomes or chromosome-arms within a cell, is a characteristic trait of human cancer [ 1 ]. Aneuploidy is associated with patient prognosis and with response to anticancer therapies [ 2 , 3 ], indicating that it can play a driving role in tumorigenesis. It is well established that the fitness advantage conferred by specific aneuploidies depends on the genomic, environmental, and developmental contexts [ 1 ]. One important cellular context is the cancer tissue of origin; aneuploidy patterns are cancer type-specific, and cancers that originate from related tissues tend to exhibit similar aneuploidy patterns [ 2 , 4 , 5 ]. Nonetheless, the selection pressures that shape the aneuploidy landscapes of human tumors are not fully understood, and it is not clear why some chromosome-arm gains and losses would recur in some tumor types but not in others.
Several non-mutually exclusive explanations have been previously provided in an attempt to explain the tissue selectivity of aneuploidy patterns. First, the densities of oncogenes (OGs) and tumor suppressor genes (TSGs) are enriched in chromosome-arms that tend to be gained or lost, respectively, potentially due to the cumulative effect of altering multiple such genes at the same time [ 6 ]. As cell proliferation is controlled in a tissue-dependent manner, the relative importance of OGs and TSGs varies across tissues, so that the density of tissue-specific driver genes can help predict aneuploidy patterns [ 7 ]. Second, some recurrent aneuploidies reflect the chromosome arm-wide gene expression patterns that characterize their normal tissue of origin, suggesting that chromosome-arm gains and losses may ‘hardwire’ pre-existing gene expression patterns [ 8 ]. Third, several strong cancer driver genes have been shown to underlie the recurrent aneuploidy of the chromosome-arms on which these genes reside; prominent examples are the tumor suppressors TP53 and PTEN , which have been shown to drive the recurrent loss of chromosome-arm 17p in leukemia and that of 10q in glioma, respectively [ 9 , 10 , 11 ]. Fourth, it has been recently proposed that somatic amplifications, including chromosome-arm gains, are positively selected in cancer evolution in order to buffer gene inactivation of haploinsufficient genes in mutation-prone regions [ 12 ].
Notably, each previous study focused on a separate aspect of tissue specificity; therefore, the relative contribution of each factor to shaping the overall aneuploidy landscape of human tumors is currently unknown. Furthermore, whether any additional tissue-specific traits could also play a major role in driving aneuploidy patterns remains an open question. Importantly, previous studies focused on the role of positive selection in driving the gain or the loss of specific chromosome-arms in specific tumor types. However, unlike point mutations in specific genes, aneuploidies come with a strong fitness cost [ 1 , 13 ]. Therefore, whereas positive selection greatly outweighs negative selection in shaping the landscape of point mutations in cancer, as evaluated by a refined version of the normalized ratio of non-synonymous to synonymous mutations [ 14 ], both positive selection and negative selection may be important for shaping the landscape of aneuploidy. Indeed, a recent study showed that negative selection could determine the boundaries of recurrent cancer copy number alterations [ 15 ]. It is therefore necessary to consider the balance between positive and negative selection in shaping the aneuploidy landscapes of human cancer.
Machine learning (ML) methods have been applied to study a variety of biological and medical questions where heterogeneous large-scale data are available [ 16 ]. In the context of cancer, supervised ML methods were applied to predict cancer driver genes [ 17 , 18 ], to distinguish between cancer types [ 19 , 20 ], and to predict gene dependency in tumors [ 21 ]. However, ML has not been applied to investigate the observed patterns of aneuploidy in human cancer. Whereas ML has been frequently used for prediction and often regarded as a black box, recent advancements have allowed more insight into the factors that underlie prediction. For example, Shapley Additive exPlanations algorithm (SHAP) [ 22 , 23 ] estimates the importance and relative contribution of each of the features utilized by the model to the model’s decisions.
Here, we present a novel ML approach to elucidate the factors that underlie the cancer type-specific patterns of aneuploidy. For this, we constructed separate ML models for chromosome-arm gain and loss, whereby each of 39 chromosome-arms within 24 cancer types was associated with 20 types of features corresponding to various genomic attributes of chromosome-arms, normal tissues, primary tumors, and cancer cell lines (CCLs). Our approach is focused on interpretation rather than prediction of aneuploidy recurrence patterns. Interpretation of the gain and loss models for aneuploidy in primary tumors captured known genomic features that had been previously reported to shape aneuploidy landscapes, supporting the models’ validity. Furthermore, these analyses suggested that negative selection played a greater role than positive selection in this process and revealed paralog compensation as an important contributor to cancer type-specific aneuploidy patterns, in both primary tumors and CCLs. Lastly, we experimentally validated a specific aneuploidy driver using genetically engineered isogenic human cells.
Constructing machine learning models to classify cancer aneuploidy patterns
To create a supervised classification ML model that predicts the recurrence pattern of aneuploidy across cancer types, we built a large‐scale dataset consisting of labels and features per instance of chromosome-arm and cancer type. For each instance, the label indicated whether the chromosome-arm was recurrently gained, lost, or remained neutral in that cancer. Labels were determined according to Genomic Identification of Significant Targets in Cancer (GISTIC2.0) [ 24 ]. We focused on 24 cancer types for which transcriptomic data of normal tissues of origin was available from the Genotype-Tissue Expression Consortium (GTEx) ([ 25 ] ( Methods ). In total, 199 instances of chromosome-arm and cancer type were labeled as gained, 307 were labeled as lost, and 430 were labeled as neutral (Fig. 1 A).
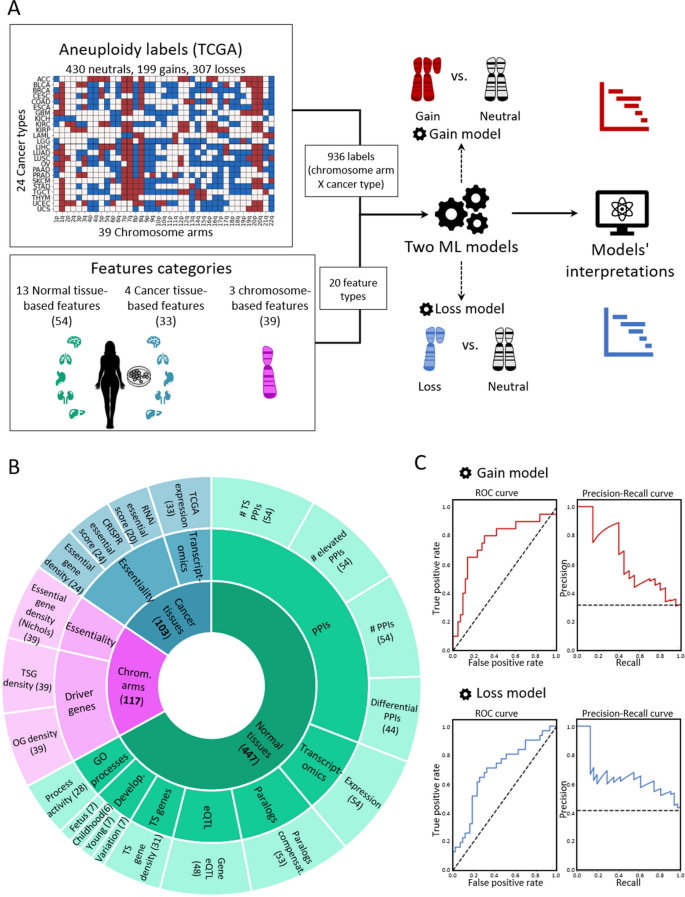
A machine learning (ML) approach for predicting aneuploidy in cancer. A Schematic view of the ML model construction. Labels represent aneuploidy status of each chromosome arm in 24 cancer types (abbreviation of cancer types detailed in Additional file 2 : Table S1), classified as gained (red, n = 199), lost (blue, n = 307), or neutral (white, n = 430). Features consist of 20 types of features pertaining to chromosome-arms, normal tissues and cancer tissues (see B ). Two separate ML models were constructed to predict gained and lost chromosome-arms (gain model and loss model). Each model was analyzed to estimate the contribution of the features to the predicted outcome. B The features analyzed by the ML model. The inner layer shows feature categories: chromosome arms (purple), cancer tissues (primary tumors and CCLs, blue), and normal tissues (green). The middle layer shows the sub-categories of the features. Chromosome-arm features include essentiality and driver genes features. Cancer-tissue features include transcriptomics and essentiality features. Normal-tissue features include protein–protein interactions (PPIs), transcriptomics, paralogs, eQTL, tissue-specific (TS) genes, development, and GO processes features. The outer layer represents all 20 feature types that were analyzed by the model. Numbers in parentheses indicate the number of tissues, organs, or cell lines from which cancer and normal tissue features were derived, or the number of chromosome-arms from which chromosome-arm features were derived. C The performance of the ML models as evaluated by the area under the receiver-operating characteristic curve (auROC, left) and the precision recall curve (auPRC, right) using tenfold cross-validation. Gain model (gradient boosting): auROC = 74% and auPRC = 63% (expected 32%). Loss model (XGBoost): auROC = 70% and auPRC = 63% (expected 42%)
Next, we defined three categories of features (Fig. 1 B; Methods ). The first category, denoted ‘chromosome-arms’, contained features of chromosome-arms that are independent of cancer type. Chromosome-arm features included the density of OGs, the density of TSGs [ 6 ], and the density of essential genes [ 26 ] per chromosome-arm. The second category, denoted ‘cancer tissues’, contained features pertaining to chromosome-arms in primary tumors and CCLs. It included features pertaining to expression of genes in primary tumors and essentiality of genes in CCLs. Expression levels of genes in each chromosome-arm per cancer type were obtained from The Cancer Genome Atlas (TCGA, https://www.cancer.gov/tcga ). Gene essentiality scores were obtained from the Cancer Dependency Map (DepMap) [ 27 ]. In total, this category included 103 omics-based readouts ( Methods ). The third category, denoted ‘normal tissues’, contained features pertaining to chromosome-arms in normal tissues from which cancer types originated (e.g., colon tissue was matched with colon adenocarcinoma, Additional file 2 : Table S1). Features of normal tissues included expression levels of genes located on each chromosome-arm in the respective normal tissue, their tissue protein–protein interactions (PPIs) [ 28 , 29 ], and their tissue-specific biological process activities [ 30 ]. It also included tissue-specific dosage relationships between paralogous genes, denoted ‘paralog compensation’ [ 31 , 32 ]. In total, this category included 447 tissue-based properties ( Methods ). To enhance our understanding of cancer and tissue selectivity, feature values of cancer and normal tissues were transformed from absolute to relative; for example, instead of indicating the absolute expression level of a gene in a given normal tissue, the expression feature was set to the expression level of the gene in the given tissue relative to its expression levels in all tissues (Additional file 1 : Fig. S1). Each chromosome-arm was then assigned with a feature value that was inferred from the values of its genes ( Methods , Additional file 1 : Fig. S2).
To fit the features dataset and the labels dataset, we further transformed the features dataset, such that each instance of chromosome-arm and cancer type was associated with features corresponding to the chromosome-arm, cancer type, and matching normal tissue ( Methods ). In total, the dataset included 20 types of features per chromosome-arm and cancer type: 3 in the chromosome-arm category, 4 in the cancer tissues category, and 13 in the normal tissues category (Fig. 1 B). We assessed the similarity between every pair of features using Spearman correlation (Additional file 1 : Fig. S3A). Most features did not correlate with each other (Additional file 1 : Fig. S3B). Among the correlated feature pairs were PPI-related features and expression in normal adult and developing tissues features (Additional file 1 : Fig. S3A). Lastly, we assessed the similarity between instances of chromosome-arm and cancer type by their feature values using principal component analysis (PCA) (Additional file 1 : Fig. S3C). Instances did not cluster by their aneuploidy pattern (gain/loss/neutral), suggesting that a more complex model is needed to classify the different patterns.
With these labels and features of each chromosome-arm and cancer type, we set out to construct two separate ML models to predict chromosome-arm gain and loss patterns across cancer types (denoted as the ‘gain model’ and the ‘loss model’, respectively; Fig. 1 A). Each model was trained and tested on data of gained (or lost) chromosome-arms versus neutral chromosome-arms. We employed five different ML methods ( Methods ) and assessed the performance of each method by using tenfold cross-validation and calculating average area under the receiver operating characteristic (auROC) and average area under the precision-recall curve (auPRC) (Additional file 1 : Fig. S4A,B). Logistic regression showed similar results to a random prediction, with auROC of 54% for each model (Additional file 1 : Fig. S4), indicating that the relationships between features and labels are non-linear. Decision tree methods that can capture such relationships [ 33 , 34 ], including gradient boosting, XGBoost, and random forest, performed better than logistic regression and similarly to each other (Additional file 1 : Fig. S4). Best performance in the gain model was achieved by gradient boosting method, with auROC of 74% and auPRC of 63% (expected: 32%) (Fig. 1 C). Best performance in the loss model was achieved by XGBoost, with auROC of 70% and auPRC of 63% (expected: 42%) (Fig. 1 C).
Revealing the top contributors to cancer aneuploidy patterns
The main purpose of our models was to identify the features that contribute the most to the recurrence patterns of aneuploidy observed in human cancer, which could illuminate the factors at play. To this aim, we used the SHAP (Shapley Additive exPlanations) algorithm [ 22 , 23 ], which estimates the importance and relative contribution of each feature to the model’s decision and ranks them accordingly. We applied SHAP separately to the gain model and to the loss model ( Methods ).
In the gain model, the topmost features were TSG density and OG density (Fig. 2 A,B). As expected, these features showed opposite directions: TSG density was low in gained chromosome-arms, whereas OG density was high, in line with previous observations [ 6 , 7 ] (Fig. 2 B). Importantly, this analysis revealed that the impact of TSGs on the gain model’s decision was twice larger than that of OGs (Fig. 2 A), highlighting the importance of negative selection for shaping cancer aneuploidy patterns. The third most important feature was TCGA expression, which quantified the expression of arm-residing genes in the given cancer type relative to their expression in other cancers. Notably, expression levels were obtained only from samples where the chromosome-arm was not gained or lost ( Methods ). This analysis revealed that, across cancer types, chromosome-arms that tend to be gained exhibit higher expression of genes even in neutral cases, consistent with a previous recent study [ 8 ]. This confirms that the genes on gained chromosome-arms are preferentially important for the specific cancer types in which these gains are recurrent. Congruently, PPIs and normal tissue expression—features of normal tissues—were also among the ten top-contributing features (Fig. 2 A). The estimated importance of all features in the gain model is shown in Additional file 1 : Fig. S5A.
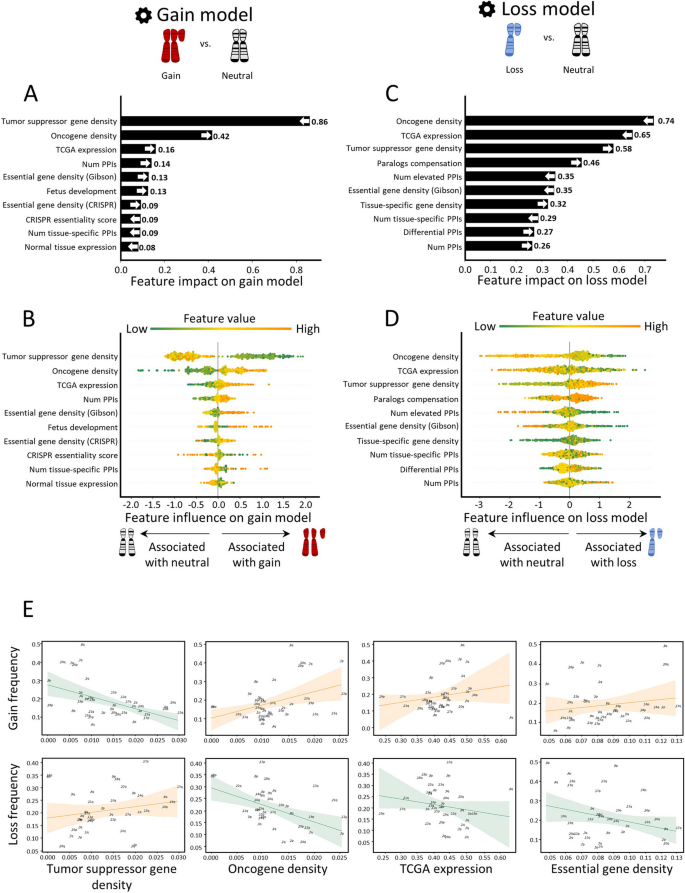
Quantitative views into the ten topmost contributing features of the gain and loss models. Features are ordered from bottom to top by their increased average absolute contribution to the model, as calculated by SHAP. A The average absolute contribution of each feature to the gain model. The directionality of the feature (i.e., whether high feature values correspond to gain or neutral) is represented by an arrow. B A detailed view of the contribution of each feature to the gain model. Per feature, each dot represents the contribution per instance of a chromosome-arm and cancer type pair. The dots are spread based on whether they were classified as neutral (left) or gain (right) by the model. Instances are colored by the feature value (green-to-orange scale denotes low-to-high value). The order (height) of each feature is the same as in A . C Same as panel A for the loss model. D Same as panel B for the loss model. E The correlations between top contributing features and the frequencies of chromosome-arm gains and losses, as measured by Spearman correlation. P -values were adjusted for multiple hypothesis testing using Benjamini–Hochberg procedure. Negative correlation between TSG density and gain frequency ( ρ = − 0.52, adjusted p = 0.006). Positive correlation between TSG density and loss frequency ( ρ = 0.3, adjusted p = 0.17). Positive correlation between OG density and gain frequency ( ρ = 0.25, adjusted p = 0.18). Negative correlation between OG density and loss frequency ( ρ = − 0.47, adjusted p = 0.01). Positive correlation between TCGA expression and gain frequency ( ρ = 0.29, adjusted p = 0.14). Negative correlation between TCGA expression and loss frequency ( ρ = − 0.33, adjusted p = 0.12). Positive correlation between essential gene density and gain frequency ( ρ = 0.16, adjusted p = 0.37). Negative correlation between essential gene density and loss frequency ( ρ = − 0.1, adjusted p = 0.5)
The loss model shared the same top three features, yet with opposite directions and different ranks (Fig. 2 C,D). OG density ranked first, was low in lost chromosome-arms, whereas TSG density ranked third, was high (Fig. 2 D), in line with previous observations [ 6 , 7 ]. In contrast to the gain model, in the loss model, the impact of OG density on the model’s decision was larger than that of TSG density, again in line with negative selection as an important force in cancer aneuploidy evolution. TCGA expression (computed from samples where the chromosome-arm was not lost or gained, see Methods ) ranked second: chromosome-arms with highly-expressed genes tended not to be recurrently lost, in line with negative selection. Another top feature that showed opposite directions between the gain and loss model was essential gene density [ 26 ]. As expected, essential gene density was low in lost chromosome-arms, in line with negative selection against losing copies of essential genes [ 26 , 27 , 35 ]. The estimated importance of all features in the loss model is shown in Additional file 1 : Fig. S5B.
To examine the direct relationships between high-ranking features and aneuploidy recurrence patterns, we assessed the correlations between these features and aneuploidy prevalence ( Methods ). In accordance with the SHAP analysis, the negative correlation between TSG density and chromosome-arm gain ( ρ = − 0.52, adjusted p = 0.0006, Spearman correlation; Fig. 2 E) was much stronger and more significant than the positive correlation between OG density and chromosome-arm gain ( ρ = 0.25, adjusted p = 0.12, Spearman correlation; Fig. 2 E). Similarly, the negative correlation between OG density and chromosome-arm loss ( ρ = − 0.47, adjusted p = 0.003, Spearman correlation; Fig. 2 E) was much stronger and more significant than the positive correlation between TSG density and chromosome-arm loss ( ρ = 0.3, adjusted p = 0.067, Spearman correlation; Fig. 2 E). TCGA expression and essential gene density were correlated with chromosome-arm gain, and anticorrelated with chromosome-arm loss, albeit to a lesser extent (Fig. 2 E, Additional file 1 : Fig. S6). Also showing positive correlations with gains and negative correlations with losses were features derived from expression levels in normal adult and developing tissues, certain PPI-related features, and additional essentiality features (Additional file 1 : Fig. S6). However, these correlations were weaker than the correlations described above. Altogether, correlation analyses supported the relationships between top features of each model and aneuploidy patterns.
The robust impact of top contributors to cancer aneuploidy patterns
Next, we asked if the above results were sensitive to our model construction schemes. We first tested the robustness of the models to internal parameters used to generate the features ( Methods ). We therefore recreated features upon modifying internal parameters and repeated model construction and interpretation ( Methods ). We found that feature importance was robust to these changes (Additional file 1 : Fig. S7, Additional file 3 : Table S2). Second, we tested the robustness of the results upon tuning the hyperparameters of each model ( Methods , Additional file 1 : Fig. S8). The top contributing features of each model were retained following hyperparameter tuning, supporting their reliability (Additional file 1 : Fig. S8C). We also checked whether the same top features would be recognized upon modeling one type of chromosome-arm event versus all other events. Applying the same approaches, we constructed two additional ML models. One model classified chromosome-arm gain versus no-gain (i.e., chromosome-arm loss or neutrality). Another model classified chromosome-arm loss versus no-loss (i.e., chromosome-arm gain or neutral). These additional models performed similarly to their respective models (Additional file 1 : Fig. S9). SHAP analysis of the two additional models revealed that feature importance was very similar between these models and the original models, which compared gained and lost chromosome-arms only to neutral chromosome-arms (Additional file 1 : Fig. S9).
We next tested whether the results were driven by a small subset of chromosome-arm and cancer type instances. For that, per model, we identified chromosome-arm and cancer type instances with the top contributions to the five topmost important features ( Methods , Additional file 4 : Table S3A,B, Additional file 5 : Table S4A,B). Most instances contributed to at least one of these features, and none of the instances contributed to all five (Additional file 5 : Table S4C). Next, we focused on chromosome-arm and cancer type instances that were top contributors to at least three of the five features (4.3% and 1.9% of the pairs in the gain and loss models, respectively). We tested their impact on the model by excluding them from the dataset and repeating the construction and interpretation of each model without them. The revised gain model retained its five topmost important features, though their ranking slightly changed (the third and fifth features switched). The revised loss model retained its four topmost important features (the fifth and seventh features switched) (Additional file 1 : Fig. S10). This suggests that the general effect of the features was not driven by a small subset of instances.
Lastly, we expanded our analyses to address whole-chromosome gains and losses. For this, we updated the features dataset to refer to whole-chromosome and cancer type instances ( Methods ). For example, the feature TSG density was updated to refer to the entire chromosome. Likewise, we updated the aneuploidy status of whole-chromosome and cancer type instances using data from GISTIC ( Methods ). This resulted in a dataset of 78 whole-chromosome gains, 151 whole-chromosome loss, and 299 neutral cases. Next, we used these data to train a whole-chromosome gain (trisomy) model and a whole-chromosome loss (monosomy) model. Model training and assessment were similar to the chromosome-arm gain and loss models. Specifically, we employed five different ML methods and assessed their performance using fivefold cross-validation. Best performance for the trisomy model was achieved by random forest, with auROC of 69% and auPRC of 47% (expected 21%; Additional file 1 : Fig. S11A). Best performance for the monosomy model was achieved by XGBoost, with auROC of 71% and auPRC of 59% (expected 34%; Additional file 1 : Fig. S11D). Performances were somewhat weaker than the chromosome-arm models, in accordance with the training data being almost twofold smaller. Lastly, we interpreted each model using SHAP. In the trisomy model, the topmost feature was TSG density and its impact was over twofold larger than the impact of other features, similarly to the chromosome-arm gain model (Additional file 1 : Fig. S11B,C). Other strong features of the chromosome-arm gain model, TCGA expression and OG density, ranked fifth and sixth, yet preserved their directionality. In the monosomy model, top features included OG density, TCGA expression, and paralogs compensation, fitting with the chromosome-arm loss model (Additional file 1 : Fig. S11E,F). The feature TSG density was ranked eight, yet preserved its directionality, similarly to the remaining features. Altogether, these results suggest that negative selection is an important factor in shaping both chromosome-arm and whole-chromosome aneuploidy patterns.
Similar features shape aneuploidy patterns in human cancer cell lines and in human tumors
Next, we aimed to test whether similar features also shape aneuploidy patterns in CCLs. We collected data of aneuploidy patterns of all chromosome-arms in CCLs [ 36 ] and analyzed 10 cancer types with matched normal tissue data from GTEx [ 25 ] ( Methods ). Similar to the analysis of cancer tissues, we labeled each instance of chromosome-arm and CCL as recurrently gained (59 instances), recurrently lost (45 instances), or neutral (286 instances) and updated the features associated with cancer types according to the CCL data ( Methods ). We then applied the gain and loss ML models, which were trained on primary tumor data, to identify determinants of aneuploidy patterns of CCLs ( Methods ). The performance of the models was at least as good as for primary tumors (gain model: auROC = 83% and auPRC = 49% (expected 15%); loss model: auROC = 76% and auPRC = 45% (expected 11%), Fig. 3 A). These results indicate that similar factors affect aneuploidy in cancers and in CCLs, consistent with the highly similar aneuploidy patterns observed in tumors and in CCLs [ 36 , 37 ].
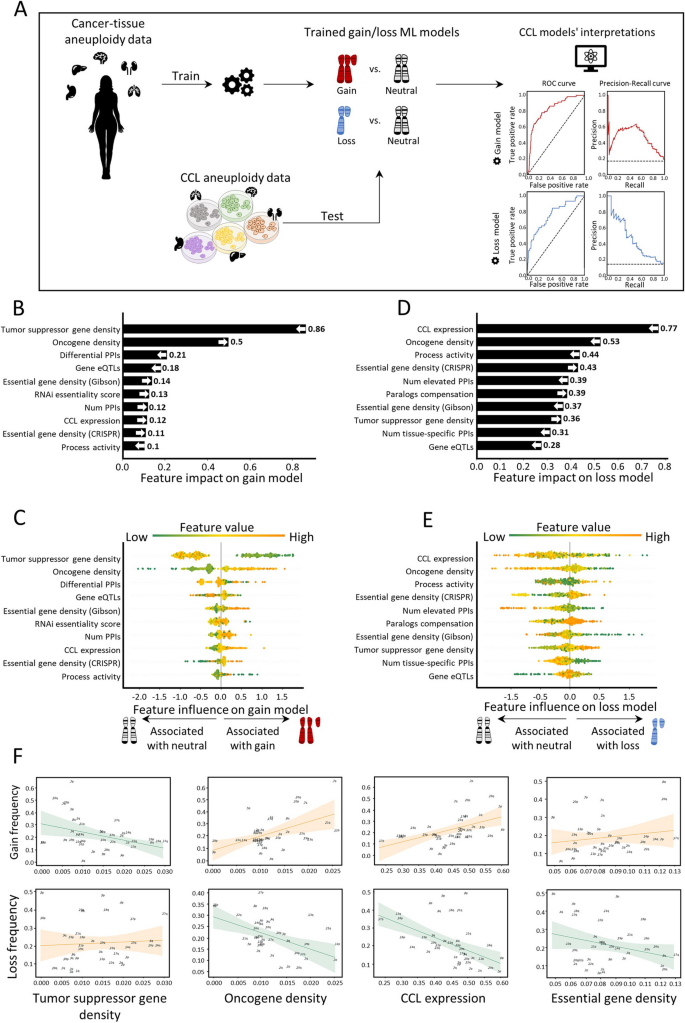
Aneuploidy patterns in CCLs and primary tumors are shaped by similar features. A The ML scheme for analysis of aneuploidy patterns in CCLs. The gain and loss models that were trained on aneuploidy patterns in primary tumors were applied to aneuploidy patterns in CCLs. Performance was measured using tenfold cross-validation. Gain model (gradient boosting): auROC = 83%, auPRC = 49% (expected 15%). Loss model (XGBoost): auROC = 76%, auPRC = 45% (expected 11%). B The average absolute contribution of the ten topmost features to the gain model (see legend of Fig. 2 A). The order and directionality of the features generally agree with the gain model in primary tumors. C A detailed view of the contribution of the ten topmost features to the gain model (see legend of Fig. 2 B). D Same as B for the loss model. The order and directionality of the features generally agree with the loss model in primary tumors. E Same as panel C for the loss model. F The correlations between top contributing features and the frequencies of chromosome-arm gains and losses, as measured by Spearman correlation. p -values were adjusted for multiple hypothesis testing using Benjamini–Hochberg procedure. Negative correlation between TSG density and gain frequency ( ρ = − 0.37, adjusted p = 0.04). Positive correlation between TSG density and loss frequency ( ρ = 0.17, adjusted p = 0.32). Positive correlation between OG density and gain frequency ( ρ = 0.44, adjusted p = 0.012). Negative correlation between OG density and loss frequency ( ρ = − 0.28, adjusted p = 0.13). Positive correlation between CCL expression and gain frequency ( ρ = 0.53, adjusted p = 0.002). Negative correlation between CCL expression and loss frequency ( ρ = − 0.6, adjusted p = 0.0006). Positive correlation between essential gene density and gain frequency ( ρ = 0.18, adjusted p = 0.33). Negative correlation between essential gene density and loss frequency ( ρ = − 0.17, adjusted p = 0.32)
We next used SHAP to assess the contribution of each feature to each of the models. TSG density and OG density remained the top contributing features for the gain model. Consistent with our results in primary tumors, the contribution of TSG density was much stronger than that of OG density, confirming the role of negative selection (Fig. 3 B,C). In the loss model, the ranking of top features was slightly different than in primary tumors (Fig. 3 D). Expression in CCL was the top feature, such that recurrently lost chromosome-arms were associated with lower gene expression in neutral cases. OG density was one of the strongest contributing features for the loss model whereas TSG density had weaker contribution, again in line with negative selection playing an important role in shaping cancer aneuploidy landscapes (Fig. 3 D,E). Certain features of normal tissues were also highly ranked. The contribution of essential gene density was also consistent with its impact in primary tumors (Fig. 3 B,C).
As with the primary tumors, correlation analyses supported the contributions of the different features. CCL expression was highly correlated with chromosome-arm gain and anticorrelated with chromosome-arm loss ( ρ = 0.54, adjusted p = 0.02, and ρ = − 0.6, adjusted p = 0.0006, respectively; Fig. 3 F). Negative correlations were also observed between TSG density and gain frequency ( ρ = − 0.37, adjusted p = 0.04, Spearman correlation; Fig. 3 F) and between OG density and loss frequency ( ρ = − 0.28, adjusted p = 0.1, Spearman correlation; Fig. 3 F). Altogether, these results indicate that despite the continuous evolution of aneuploidy throughout CCL culture propagation [ 38 ], similar features drive aneuploidy recurrence patterns in primary tumors and in CCLs.
Chromosome 13q aneuploidy patterns are tissue-specific, and KLF5 is a driver of 13q gain in colorectal cancer
In human cancer, a chromosome-arm is either recurrently gained across cancer types or it is recurrently lost across cancer types, but rarely is a chromosome-arm both gained in some cancer types and lost in others [ 4 , 5 ]. An intriguing exception is chr13q. Of all chromosome-arms, chr13q is the chromosome-arm with the highest density of tumor suppressor genes (Fig. 2 E). It is therefore not surprising that chr13q is recurrently lost across multiple cancer types (with a median of 30% of the tumors losing one copy of 13q across cancer types) [ 4 , 5 ]. Interestingly, however, chr13q is recurrently gained in human colorectal cancer (in 58% of the samples), suggesting that it can confer a selection advantage to colorectal cells in a tissue-specific manner. Indeed, when comparing colorectal tumors and colorectal cancer cell lines against all other cancer types, chr13q was the top differentially affected chromosome-arm (Fig. 4 A,B). We therefore set out to study the basis for this unique tissue-specific aneuploidy pattern.
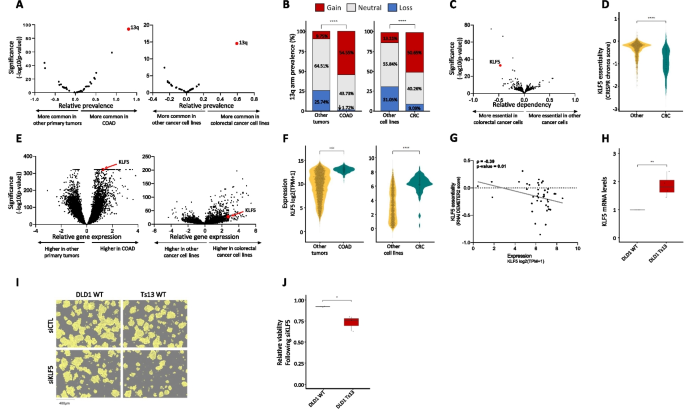
KLF5 is a potential driver of chromosome 13q gain in human colorectal cancer. A Comparison of the prevalence of chromosome-arm aneuploidies in colorectal tumors against all other tumors (left) and colorectal cancer cell lines against all other cancer cell lines (right). On the right side are the aneuploidies that are more common in colorectal cancer, and on the left side are the ones that are less common in colorectal cancer. Chromosome-arm 13q (in red) is the top differential aneuploidy in colorectal cancer. B Comparison of the prevalence of 13q aneuploidy between colorectal tumors and all other tumors (left) and between colorectal cancer cell lines and all other cancer cell lines (right). ****, p < 0.0001 and ****, p < 0.0001; Chi-square test. C Genome-wide comparison of differentially essential genes between colorectal cancer cell lines ( n = 85) and all other cancer cell lines ( n = 1407). On the right side are the genes that are more essential in other cancer cell lines, and on the left side are those that are more essential in colorectal cancer, based on a genome-wide CRISPR/Cas9 knockout screens [ 39 ]. The x -axis presents the effect size (i.e., the differential response between colorectal cell lines and other cell lines), and the y -axis presents the significance of the difference (-log10( p -value)). KLF5 (in red) is the second most differentially essential gene in colorectal cancer cell lines. D Comparison of the sensitivity to CRISPR knockout of KLF5 between colorectal cancer cell lines ( n = 59) and all other cancer cell lines ( n = 1041). ****, p < 0.0001; two-tailed Mann–Whitney test. E Genome-wide comparison of differentially expressed genes between colorectal tumors ( n = 434) and all other tumors (on the left, n = 11,060) and between colorectal cancer cell lines ( n = 85) and all other cancer cell lines (on the right, n = 1407). On the right side are the genes that are over-expressed in colorectal cancer and on the left side are those that are over-expressed in other cell lines. KLF5 (in red) significantly over-expressed in colorectal cancer. F Comparison of KLF5 mRNA levels between colorectal tumors ( n = 434) and all other tumors on the left ( n = 11,060) and between colorectal cancer cell lines ( n = 85) and all other cancer cell lines (on the right, n = 1407). ****, p < 0.0001; two-tailed Mann–Whitney test. G Correlation between KLF5 mRNA expression and the sensitivity to KLF5 knockdown, showing that higher KLF5 expression is associated with increased sensitivity to its RNAi-mediated knockdown. ρ = − 0.39, p = 0.01; Spearman correlation. H Comparison of KLF5 mRNA levels between DLD1-WT (without trisomy of chromosome 13) and DLD1-Ts13 (with trisomy of chromosome 13) colorectal cancer cells. **, p = 0.0025; one-sample t -test. I Representative images of DLD1-WT and DLD1-Ts13 cells treated with siRNA against KLF5 . DLD1-Ts13 cells proliferated more slowly, as previously reported, but were more sensitive to the knockdown after accounting for their basal proliferation rate. Cell masking (shown in yellow) was performed using live cell imaging (IncuCyte) following 72 h of treatment. Scale bar 400µm. J Quantification of the relative response to KLF5 knockdown between DLD1-WT and DLD1-Ts13, as evaluated by quantifying cell viability in cells treated with siRNA against KLF5 versus a control siRNA for 72 h. n = 3 independent experiments. *, p = 0.0346; one-sided paired t -test
We performed a genome-wide comparison of differentially essential genes between colorectal cell lines and all other cell lines. The two top genes, which are much more essential in colorectal cancer cells than in other cancer types, were CTNNB1 and KLF5 (Fig. 4 C). Of particular interest is KLF5 , which is located on chr13q and colorectal cancer cell lines are significantly more sensitive to its knockout (Fig. 4 D). KLF5 was reported to be tumor-suppressive in the context of several cancer types, such as breast and prostate [ 40 , 41 ]. In colon cancer, however, not only is KLF5 important for tissue identity [ 42 ], but it was also reported to be haploinsufficient [ 43 ], potentially explaining why loss of chr13q is so rare in colorectal cancer. In line with a potential driving role in the recurrence of chr13q gain in colorectal cancer, KLF5 was among the most significantly overexpressed genes in colorectal tumors and in colorectal cell lines versus all other cancer types (Fig. 4 E,F). Furthermore, KLF5 expression levels correlated with the cells’ sensitivity to its knockdown (Fig. 4 G). To confirm the association between chr13q gain and KLF5 expression and dependency, we next turned to an isogenic system of human colon cancer cells (DLD1) into which trisomy 13 had been introduced (DLD1-Ts13) [ 44 ]. Using this unique experimental system, we confirmed that trisomy 13 results in overexpression of KLF5 (Fig. 4 H) and increased sensitivity to its siRNA-mediated genetic depletion (Fig. 4 I,J and Additional file 1 : Fig. S12, Additional file 1 : Fig. S13). This differential response was specific to KLF5 , as the trisomy did not affect the sensitivity of the cells to a control siRNA (Additional file 1 : Fig. S14), to knockdown of an unrelated gene residing on chr13q ( NEK3 ; Additional file 1 : Fig. S15), or to knockdown of another transcription factor that plays a role in colon development and is located on another chromosome ( TTC7A , located on chr2p; Additional file 1 : Fig. S16). We, therefore, propose that KLF5 contributes to the uniquely variable pattern of chr13q aneuploidy across cancer types.
Paralog compensation is an important feature shaping tissue-specific aneuploidy patterns
One of the topmost contributing features to the chromosome-arm loss model in primary tumors and in CCLs, as well as to the whole-chromosome loss model, was paralog compensation. It was previously shown that while loss of genes with paralogs was less detrimental than loss of singleton genes [ 45 ], the impact of gene loss in a specific condition depends on the expression level of its paralog [ 46 ]. The paralog compensation feature was therefore designed to quantify the expression ratio between two paralogs. Specifically, higher values of this feature for a given gene correspond to a higher expression of the paralog relative to the gene ( Methods ). Previous studies of hereditary disease genes showed that lower paralog compensation in a tissue was associated with disease manifestation in that tissue [ 31 , 32 ]. Paralog compensation was also shown in cancer tissues: In CCLs, essentiality of a gene was decreased with an increased expression of its paralog [ 27 , 46 , 47 ]. In primary tumors, paralog compensation was shown to be associated with increased prevalence of non-synonymous mutations [ 48 ] and to correlate with the prevalence of homozygous gene deletion [ 49 ]. However, the contribution of paralog compensation to aneuploidy has not been studied to date.
Paralog compensation ranked fourth and sixth in the loss models of primary tumors and CCLs, respectively (Fig. 2 C, Fig. 3 D). In both, chromosome-arm loss was associated with higher paralog compensation, suggesting that loss is facilitated by higher relative expression of paralogs (Fig. 2 D, Fig. 3 E). We also analyzed the correlations between the frequency of chromosome-arm loss and paralog compensation ( Methods , Fig. 5 A). Indeed, the frequency of chromosome-arm loss was positively correlated with paralog compensation in both primary tumors and in CCLs ( ρ = 0.26 and ρ = 0.46, respectively, Spearman correlation; Fig. 5 A).
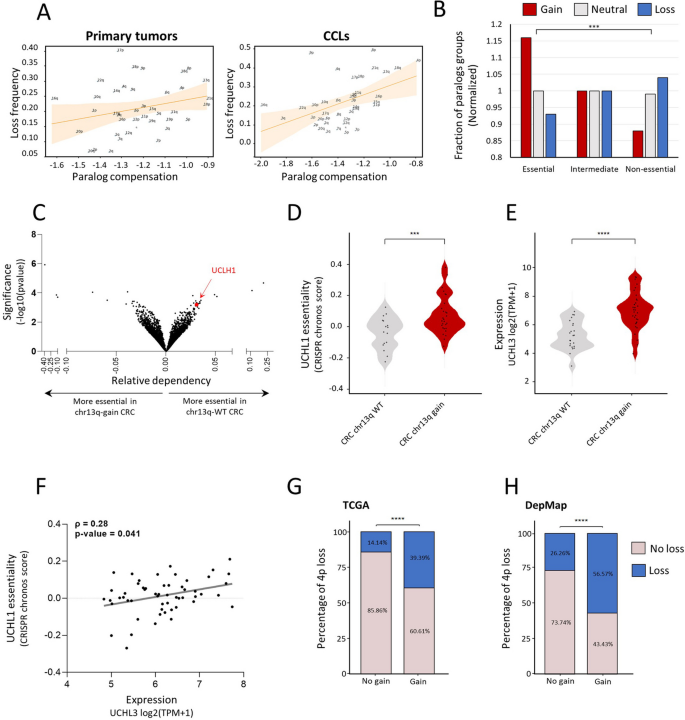
Paralog compensation is an important feature shaping tissue-specific aneuploidy patterns. A The correlation between paralog compensation values and loss frequency of chromosome arms in primary tumors (left, ρ = 0.26, adjusted p = 0.18, Spearman correlation) and in CCLs (right, ρ = 0.46, adjusted p = 0.01, Spearman correlation). B A view into the aneuploidy patterns of paralogs of recurrently lost genes. Recurrently lost genes were divided into essential, intermediate, and non-essential groups. Paralogs of essential genes were more frequently gained, whereas paralogs of non-essential genes were more frequently lost. C Genome-wide comparison of differentially essential genes in colorectal cell lines with chr13q gain ( n = 39) versus chr13q-WT colorectal cell lines ( n = 25). On the right side are the genes that are more essential in chr13q-WT cells, and on the left side those that are more essential in chr13q-gain cells, based on a genome-wide CRISPR/Cas9 knockout screens [ 39 ]. The x -axis presents the effect size (i.e., the differential response between chr1q-WT and chr13q-gain colorectal cell lines) and the y -axis presents the significance of the difference (-log10(p-value)). UCHL1 (in red) is one of the top genes identified to be more essential in chr13q-WT cells. D Comparison of the sensitivity to CRISPR knockout of UCHL1 between colorectal cell lines with ( n = 28) and without chr13q gain ( n = 16). ***, p = 0.0003; two-tailed Mann–Whitney test. E Comparison of UCHL3 mRNA expression between colorectal cell lines with ( n = 34) and without chr13q gain ( n = 23). ****, p < 0.0001; two-tailed Mann–Whitney test. F Correlation between UCHL3 mRNA expression and the sensitivity to UCHL1 knockout, showing that higher UCHL3 mRNA levels are associated with reduced sensitivity to UCHL1 knockout. ρ = 0.28, p = 0.041; Spearman correlation. G Comparison of the prevalence of chr4p loss between human primary colorectal tumors with and without chr13q gain. ****, p < 0.0001, Chi-square test. H Comparison of the prevalence of chr4p loss between human colorectal cancer cell lines with and without chr13q gain. ****, p < 0.0001, Chi-square test
Next, we tested whether paralog compensation, namely gain or overexpression of paralogs, could indeed facilitate chromosome-arm loss. We started by grouping genes in recurrently lost chromosome-arms into essential, intermediate, or non-essential, according to their essentiality in CCLs [ 27 ] ( Methods ). We then associated each gene with the aneuploidy status of the chromosome-arm of its paralog, namely whether the chromosome-arm of the paralog was gained, lost, or remained neutral in the corresponding CCL ( Methods , Additional file 1 : Fig. S17A). The fraction of genes with paralogs on neutral chromosome-arms was similar in all essentiality groups (Fig. 5 B). In contrast, the fraction of gained paralogs was highest in the group of essential genes and lowest in the group of non-essential genes. This suggests that the loss of essential genes is more likely accompanied by the gain of their paralogs. Likewise, the fraction of lost paralogs was lowest in the group of essential genes and highest in the group of non-essential genes ( p = 2.38e − 24, Chi-square test; Fig. 5 B). This suggests that the loss of essential genes is less likely to be accompanied by the loss of their paralog. The same trend was shown upon comparing the distribution of essentiality scores between genes with gained paralogs versus genes with lost paralogs ( p = 9.2e − 16, KS test; Additional file 1 : Fig. S17B). Hence, paralog compensation can facilitate chromosome-arm loss.
Next, we decided to identify a specific example. In human colon cancer, the long arm of chromosome 13 (chr13q) is commonly gained, as described above, whereas the short arm of chromosome 4 (chr4p) is commonly lost [ 5 , 37 ]. We analyzed the association between chr13q-residing genes and the essentiality of their paralogs, revealing UCHL3 (chr13q)- UCHL1 (chr 4p) as the most significant correlation (Additional file 6 : Table S5 and Fig. 5 C). Human colon cancer cell lines with chr13q gain were less sensitive to CRISPR/Cas9-mediated knockout of UCHL1 (Fig. 5 D). Consistently, chr13q-gained cell lines had significantly higher mRNA levels of UCHL3 (Fig. 5 E), and the expression of UCHL3 was significantly correlated with the essentiality of UCHL1 (Fig. 5 F). We hypothesized that the relationship between these paralogs may affect the co-occurrence patterns of the chromosome-arms on which they reside. Indeed, both in primary human colon cancer and in colon cancer cell lines, loss of chr4p was significantly more prevalent when chr13q was gained (Fig. 5 G,H). Together, these results demonstrate that paralog compensation can be affected by—and contribute to the shaping of—aneuploidy patterns.
Recurrent aneuploidy patterns are an intriguing phenomenon that is only partly understood. Several previous studies characterized the unique patterns of aneuploidy in cancer [ 4 , 5 , 50 ] or attempted to identify the driving role of a specific aberration in a specific cancer context [ 9 , 51 , 52 , 53 , 54 ]. Attempts to explain copy number patterns in cancer focused on specific pre-defined aspects, such as the specific boundaries of the alterations [ 15 ], the densities of OGs and TSGs on the aberrant chromosomes [ 6 , 7 ] or the gene expression changes that they induce [ 8 ], and these aspects were interrogated using statistical methods and correlation analyses. Here, in contrast, we studied this phenomenon using an unbiased ML-based approach. As with other ML applications, it allowed us to study multiple aspects simultaneously. Yet, unlike classical ML-based studies that mainly aim to improve prediction, for example by using deep learning to predict gene dependency in tumors [ 21 ], our focus was on interpretability. In fact, we built chromosome-arm gains and loss models only to then identify factors that shape aneuploidy patterns. Interpretable ML was recently applied to reveal genetic attributes that contribute to the manifestation of Mendelian diseases [ 55 ]. In this study, we applied interpretable ML for the first time in the context of aneuploidy and at chromosome-arm resolution.
The capability of ML to concurrently assess multiple features opened the door for assessing the relevance of features that have not been rigorously studied to date, such as paralog compensation. Yet, ML has its limitations. Mainly, the number of features that could be analyzed depends on the size of the labeled dataset [ 56 ], which, in aneuploidy, was restricted by the number of chromosome-arms and cancer types. We therefore analyzed 20 types of features and tested linear regression and tree-based ML methods, which, unlike deep learning, are suitable for this size of data. Following prediction, our main goal was to assess the relative contribution of each feature to the model’s decision and its directionality using SHAP. Nevertheless, SHAP results should be interpreted with caution. First, SHAP assumes feature independence, although features could be correlated with each other or confounded. Importantly, we found that only a small subset of features correlated with each other, and they did not include the topmost contributing features (Additional file 1 : Fig. S3A). Second, the top contributing factors could be correlated with prediction strength, rather than being causal. Lastly, due to the hierarchical nature of decision trees, features that are located low in the decision tree explain only a small fraction of the cases. To estimate feature contribution and directionality more broadly, we explicitly correlated feature values with chromosome-arm gain and loss frequency, finding support for their broad relevance (Fig. 2 E, Additional file 1 : Fig. S6). We also conducted multiple analyses that tested the robustness of the results to the models’ construction schemes (Additional file 1 : Fig. S7, S8), the modeled events (one event versus rest, Additional file 1 : Fig. S9; whole-chromosome, Additional file 1 : Fig. S11), or to a subset of the chromosome-arm and cancer type instances (Additional file 1 : Fig. S10). The different analyses repeatedly revealed the same factors at play, supporting the reliability of our results.
The features that we studied included known and previously underexplored attributes of chromosome-arms, healthy tissues and cancer cells (Fig. 1 A,B). OG and TSG densities, which have previously been observed to be enriched on gained and lost chromosome-arms, respectively [ 6 , 7 ], were top contributing features in both models, thereby supporting the validity of our approach (Fig. 2 A,C). In the gain model in particular, their contribution was over 2.6 and 5 times stronger, respectively, than any other feature (Fig. 2 A). As our TSG and OG features were cancer-independent, their importance may explain the observation that certain chromosome-arms tend to be either gained or lost across multiple cancer types [ 4 , 5 ]. Their relative contribution, however, was surprising. In both models, negative associations were much stronger than positive associations: OG density contributed to chromosome-arm loss more than TSG density, implying that it was more important to maintain OGs than to lose TSG (Fig. 2 B,D). The reciprocal relationship was true for chromosome-arm gain, as it was more important to maintain TSGs than to gain OGs (Fig. 2 A,C). These results were validated using correlation analyses (Fig. 2 E) and were recapitulated in CCLs (Fig. 3 ) and in the analysis of whole-chromosome gains and losses (Additional file 1 : Fig. S11). Together, they highlight the importance of negative selection for shaping cancer aneuploidy landscapes [ 1 , 15 ].
A known factor that contributed to both models was gene expression in primary tumors (TCGA expression, Fig. 2 ) and in CCLs (CCL expression, Fig. 3 ). This result suggests that cancers tend to gain chromosome-arms that are enriched for highly-expressed genes and tend to lose chromosome-arms that are enriched for lowly expressed genes. A Similar trend was shown recently for gene expression in normal tissues [ 8 ]. Our approach was capable of comparing the relative contributions of both features. We found that the contribution of gene expression in normal tissue was lower than that in cancer tissues, as also evident by its lower correlation with the frequencies of chromosome-arm gains and losses (Additional file 1 : Fig. S6). Nevertheless, other features that were derived from gene expression in normal tissues ranked highly, such as the number of PPIs in the gain model and paralog compensation in the loss model, and hence expression in normal tissues is also important (Fig. 2 ).
A previously under-explored feature that we considered was paralog compensation. Paralog compensation was shown to play a role in the manifestation of Mendelian and complex diseases [ 31 , 32 ] and in the dispensability of genes in tumors [ 48 , 49 ] and CCLs [ 27 , 46 , 47 ], but was not studied in the context of aneuploidy. Here, paralog compensation was among the top contributors to the loss model (Fig. 2 C, Fig. 3 D). The directionality of this feature and correlation analyses showed that, relative to genes located on neutral chromosome-arms, genes located on lost chromosome-arms tend to have higher compensation by paralogs (Fig. 5 A). This suggests that chromosome-arm loss is facilitated, or better tolerated, through paralogs’ expression. We also showed that the more essential recurrently lost genes are, the more likely they are to be associated with gains of paralog-bearing chromosome-arms (Fig. 5 B). We further demonstrated this for a specific example (the UCHL3 - UCHL1 paralog pair; Fig. 5 ). Overall, our analysis reveals that compensation between paralogs through expression or chromosome-arm gain plays an important role in shaping the landscape of chromosome-arm loss.
Combining the different results, our models reveal a previously under-appreciated role for negative selection in driving human cancer aneuploidy. This was evident by the tendency not to lose chromosome arms with high OG density, high frequency of essential genes, or low compensation by paralogs, and not to gain chromosome arms with high TSG density (Fig. 6 ). Previous studies have shown that positive selection outweighs negative selection in shaping the point mutation landscape of human tumors [ 14 ]. However, the strong fitness cost associated with aneuploidy suggests that the aneuploidy landscape of tumors might be strongly affected by negative selection as well (reviewed in [ 1 ]). Interestingly, evidence for the involvement of negative selection in shaping the copy number alteration (CNA) landscapes of tumors has been proposed in a recent study that analyzed CNA length distributions across human tumors [ 15 ]. Our study thus lends further independent support to the importance of negative selection in shaping the landscape of aneuploidy across human cancers (Fig. 6 ).
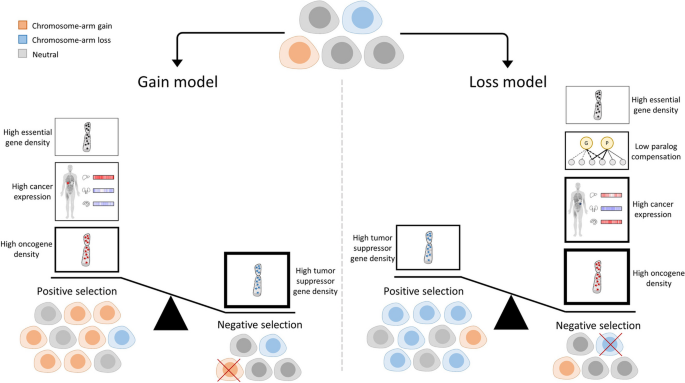
A schematic presentation of the results of the study. Cancer evolution is shaped by negative and positive selection leading to enrichment or depletion of cells with distinct aneuploidy patterns. In the gain model (left), main contributors to positive selection of gained chromosome arms are: (1) high oncogene density, (2) high expression of genes in the cancer tissue, and (3) high essential gene density. A major contributor to negative selection is high tumor suppressor gene density. Importantly, the density of TSGs is more important than the density of OGs for predicting chromosome-arm gains. In the loss model (right), a main contributor to positive selection of lost chromosome arms is high tumor suppressor gene density. Major contributors to negative selection are high oncogene density, high expression of genes in the cancer tissue, low compensation by paralogs, and high density of essential genes. In both models, the features associated with negative selection have higher overall contribution than features associated with positive selection. The thickness of the borders of the boxes reflects the relative contribution of the features to the model
Our genome-wide analysis could be expanded in future studies in several ways: (1) While we focused on the top-contributing features, other features, such as PPIs that contributed to both gain and loss models, are also relevant and remain to be studied in depth. (2) It will be interesting to consider additional types of aneuploidy, such as tetrasomies, and explore how whole-genome doubling affects the importance of the features in shaping the aneuploidy landscapes of tumors. (3) Tumors often exhibit heterogeneous (mosaic) aneuploidy patterns [ 57 , 58 , 59 , 60 ]. Our analyses were entirely based on bulk-population data, and our results therefore describe the selection pressures that shape the landscape of clonal aneuploidies. As more single-cell omics data becomes available, it will be interesting to also study the selection pressures that shape subclonal aneuploidy patterns. (4) Aneuploidies do not always arise independently, so that chromosome-arm events can co-occur or be mutually exclusive [ 37 ]. We show that only a small fraction of chromosome-arm events co-occur (Additional file 7 : Table S6), suggesting that their effect on our models would likely be small. Nonetheless, considering co-occurrence patterns could further refine the models.
Lastly, we explored one example of a unique aneuploidy pattern (chr13q) that is recurrently altered in opposite directions in different cancer types. In line with tumor suppressors and oncogenes being a major feature explaining aneuploidy patterns, we identified KLF5 as a colorectal-specific dependency gene. Using an isogenic system of colorectal cancer cells with/without gain of chr13, we experimentally demonstrated that this aneuploidy is associated with increased expression and increased essentiality of KLF5 . The finding that colorectal cells with trisomy 13 are more sensitive to KLF5 depletion suggests positive selection for its gain, on top of a potential negative selection against a deleterious loss. We therefore propose that KLF5 might explain why chr13q is commonly gained and rarely lost in colorectal cancer, unlike its recurrent loss across multiple other cancer types.
Overall, our study provides novel insights into the forces that shape the tissue-specific patterns of aneuploidy observed in human cancer and demonstrates the value of applying ML approaches to dissect this complicated question. Our results suggest that aneuploidy patterns are shaped by a combination of tissue-specific and non-tissue-specific factors. Negative selection in general and paralog compensation in particular play a major role in shaping the aneuploidy landscapes of human cancer and should therefore be computationally modeled and experimentally studied in the research of cancer aneuploidy.
Chromosome-arm aneuploidy patterns per cancer
Chromosome-arm events per cancer were defined according to GISTIC2.0 [ 24 ] for all (39) chromosome-arms in 24 cancer types for which data of the normal tissue of origin was available from GTEx [ 25 ]. GISTIC2.0 computed the probability of chromosome-arm events by comparing the observed frequency to the expected rate, while considering chromosome-arm length and other parameters [ 61 ]. A chromosome-arm was considered as gained or lost in a specific cancer if the q -value of its amplification or deletion, respectively, was lower than 0.05. Otherwise, the chromosome-arm was considered as neutral. In case the q -value of both amplification and deletion was lower than 0.05, decision was made based on the lower q -value. In case of a tie, the more frequent event was selected. GISTIC2.0 data, including q -values and frequencies, were downloaded from ref. [ 62 ]. Lastly, we analyzed co-incidence probabilities of chromosome-arm events per cancer. Co-incidence probabilities for chromosome-arms and cancers in our dataset were obtained from [ 37 ].The median fraction of chromosome-arm pairs with significant co-incidence per cancer was 2.05% (Additional file 7 : Table S6). Hence, the impact of co-incidence on the models is expected to be small.
We also carried separate analyses of gain and loss of whole-chromosomes. A whole-chromosome was considered as gained if the q -value of the amplification of its two arms was lower than 0.05. Likewise, a whole-chromosome was considered as lost if the q -value of the deletion of its two arms was lower than 0.05.
Construction of a features dataset of instances of chromosome-arm and cancer type pairs
For each chromosome-arm and cancer, we created features that were inferred from data of chromosome-arms, genes, cancer tissues and CCLs, and normal tissues (Fig. 1 B, Additional file 2 : Table S1). A schematic pipeline of the dataset construction appears in Additional file 1 : Fig. S1. The different types of features are described below.
Features of chromosome-arms
Each chromosome-arm was associated with three types of features, including oncogene density, tumor suppressor gene density, and essential gene density. Oncogene density and tumor suppressor gene density per chromosome-arm were obtained from Davoli et al. [ 6 ]. Data of essential genes was obtained from Nichols et al. [ 26 ], where a gene was considered essential if its essentiality probability was > 0.8. The density of essential genes per chromosome-arm was calculated as the fraction of essential genes out of the protein-coding genes on that chromosome-arm. Next, we associated each instance of chromosome-arm and cancer type with features of that chromosome-arm.
Features of cancer tissues
Each instance of chromosome-arm and cancer type was associated with four types of cancer-related features, including transcriptomics, essentiality by CRISPR or RNAi in CCLs, and cancer-specific density of essential genes. Transcriptomics was based on transcriptomic profiles of 33 cancer types from TCGA [ 63 ] that were obtained from GDC Xena Hub v18.0 (updated 2019–08-28). Per cancer, we associated each gene with its median expression level in samples of that cancer. To avoid expression bias due to chromosome-arm gain or loss, the median expression of each gene was computed from samples where the chromosome-arm harboring the gene was neutral according to Taylor et al. [ 5 ]. Essentiality by CRISPR was based on CRISPR screens of 24 CCLs from the DepMap portal version 21Q1. Essentiality by RNAi was based on RNAi data of 20 CCLs from DepMap [ 27 ]. In each of these datasets, the score of each gene indicated the change, relative to control, in the growth rate of the cell line upon gene inactivation via CRISPR or RNAi. Accordingly, genes with negative scores were essential for the growth of the respective cell line. We associated each gene with its median essentiality score based on either CRISPR or RNAi per cell line. To reflect gene essentiality more intuitively, we reversed the direction of the scores (multiplied them by − 1), so that more essential genes had higher scores. To avoid bias due to chromosome-arm gain or loss, the median essentially of each gene was computed from samples where the chromosome-arm harboring the gene was neutral [ 5 ]. Cancer-specific density of essential genes was calculated as the fraction of essential genes (CRISPR-based essentiality score > 0.5) in a given CCL out of the protein-coding genes residing on that chromosome-arm.
Features of normal tissues
Each instance of chromosome-arm and cancer type was associated with 13 types of features that were derived from [ 55 ]. We associated each cancer type with the normal tissue in which it originates (Additional file 2 : Table S1).
Transcriptomics
Data of normal tissues included transcriptomic profiles of 54 adult human tissues measured via RNA-sequencing from GTEx v8 [ 25 ]. Each gene was associated with its median expression in each adult human tissue. Genes with median TPM > 1 in a tissue were considered as expressed in that tissue.
Tissue-specific genes
Per gene, we measured its expression in a given tissue relative to other tissues using z -score calculation. Genes with z -score > 2 were considered tissue-specific. Lastly, we associated each chromosome-arm and tissue with the density of tissue-specific genes.
PPI features
Each gene was associated with the set of its PPI partners. We included only partners with experimentally detected interactions that were obtained from MyProteinNet web-tool [ 64 ]. Per each tissue, we associated each gene with four PPI-related features:
“Number PPIs” was set to the number of PPI partners that were expressed in that tissue.
“Number elevated PPIs” relied on preferential expression scores computed according to [ 28 ] and was set to the number of PPI partners that were preferentially expressed in that tissue (preferential expression > 2, [ 65 ].
“Number tissue-specific PPIs” was set to the number of PPI partners that were expressed in that tissue and in at most 20% of the tissues.
“Differential PPIs” relied on differential PPI scores per tissue from The DifferentialNet Database [ 28 ] and was set to gene’s median differential PPI score per tissue. If the gene was not expressed in a given tissue, its feature values in that tissue were set to 0.
Differential process activity features
Differential process activity scores per gene and tissue were obtained from [ 30 ]. The score of a gene in a given tissue was set to the median differential activity of the Gene Ontology (GO) processes involving that gene. The differential activity was relative to the activity of the same processes in other tissues.
eQTL features
eQTLs per gene and tissue were obtained from GTEx [ 25 ]. Each gene was associated with the p -value its eGene in that tissue.
Paralog compensation features
Each gene was associated with its best matching paralog according to Ensembl-BioMart. Per tissue, the gene score was set to the median expression ratio of the gene and its paralog, as described in [ 31 , 32 ]. Accordingly, high values mark genes with low paralog compensation.
Development features
Transcriptomic data of seven human organs measured at several time points during development were obtained from [ 66 ]. We united time points into time periods including fetal (4–20 weeks post-conception), childhood (newborn, infant, and toddler), and young (school, teenager and young adult). Per organ, we associated each gene with its median expression level per period. Next, we created an additional feature that reflected the expression variability of each gene across periods.
Transforming gene features into chromosome-arm features
Some of the features described above referred to genes. To create chromosome-arm-based features, we grouped together genes that were located on the same chromosome-arm [ 67 ]. Next, to highlight differences between tissues, for each feature, we associated a gene with its value in that tissue relative to other tissues. Features that were already tissue-relative, including “Differential PPIs” and “Differential process activity,” were maintained. Other features were converted into tissue-relative values via a z -score calculation (see Eq. 1 ). Lastly, per feature, we ranked genes by their tissue-relative score and associated each chromosome-arm with the median score of the genes ranking at the top 10% (Additional file 1 : Fig. S2). Transcriptomic features in the testis and whole blood were highly distinct from other tissues; we normalized all transcriptomic features per tissue. To reflect paralog compensation more intuitively, we reversed the direction of the resulting features (multiplied them by − 1), so that genes with higher compensation had higher scores.
T denotes the set of tissues, G denotes the set of genes, v denotes the value of the feature, and σ denotes the standard deviation.
Construction of the final dataset
The features described above referred to chromosome-arms in cancers, CCLs, and normal tissues. To create chromosome-arm features per cancer, we associated each cancer with the chromosome-arm features of its tissue of origin and CCL (Additional file 2 : Table S1). For features of normal tissues where multiple sub-regions were sampled (e.g., skin sun-exposed and not sun-exposed, or brain sub-regions), we set the chromosome-arm values to their median across sub-regions. The final dataset contained features for all 936 instances of 39 chromosome-arms and 24 cancers for which the cancer’s normal tissue of origin was available in GTEx [ 25 ] (Additional file 2 : Table S1). We assessed the similarity between every pair of features using Spearman correlation (Additional file 1 : Fig. S3A). We assessed whether chromosome-arm and cancer type instances had similar feature values using PCA (Additional file 1 : Fig. S3C).
ML application to model chromosome-arm and cancer aneuploidy
Below we describe the ML method used for aneuploidy classification and the SHAP (SHapley Additive exPlanations) analysis of feature importance that was used to interpret the resulting models.
Aneuploidy ML classification models
We constructed two ML models: a gain model that compared between gained and unchanged (neutral) chromosome-arms and a loss model that compared between lost and unchanged (neutral) chromosome-arm.
ML comparison and implementation
Per model, we tested several ML methods, including logistic regression, XGBoost, gradient boosting, random forest, and bagging. All ML methods were implemented using the Scikit-learn python package [ 68 ], except for XGB, which was implemented using the Scikit-learn API of the XGBoost package [ 69 ]. To assess the performance of each model, we used tenfold cross-validation. Then, we calculated the au-ROC and the au-PRC. Each point on the curve corresponded to a particular cutoff that represented a trade-off between sensitivity and specificity and between precision and recall, respectively.
SHAP analysis of feature importance
To measure the contribution and importance of the different features, we used SHAP algorithm [ 70 ]. SHAP is a game-theoretic approach to explain the output of ML models: for each feature, SHAP assigns a contribution value to each instance of chromosome-arm and cancer type. It then estimates the contribution of that feature to the model by the average absolute SHAP values of all instances. Per model, we created the SHAP plots corresponding to feature contribution and directionality. In both, features were ordered by their importance to the model (top meaning most contributing). We also visualized the directionality of each feature using arrows in the SHAP bar plot. The direction of the arrow showed whether the highest values of that feature (top 50%) corresponded to a chromosome-arm event (gain or loss, right) or to neutrality (left).
Robustness analyses
We analyzed the robustness of the models and their interpretation with respect to internal parameters used to generate the features and the hyperparameters of the ML models. For feature generation, we used top 10% of genes with highest values to calculate each gene-based chromosome-arm feature. We therefore reconstructed features by also using the top 1%, 5%, 15%, and 20% of the genes. We then assessed the performance of each method using tenfold cross-validation. In all cases, method performance was similar (Additional file 3 : Table S2). SHAP analysis of the best performing method per case showed similar results with respect to the topmost contributing features and their directionality (Additional file 1 : Fig. S7). For robustness to parameter choices, we tuned the hyperparameters per ML method separately for the gain model and for the loss model, and repeated model construction and interpretation. Tuning was optimized for precision and performed using the “RandomizedSearch” function of sklearn python package, with number of sampled parameters (iterations, n_iter) set to 200 and tenfold cross-validation. Best parameters per method and model and their performance appear in Additional file 1 : Fig. S8A,B. Performance was only slightly improved, and interpretation of the best performing models revealed similar results (Additional file 1 : Fig. S8C).
Lastly, we tested if the most important features per model were driven by a small subset of chromosome-arm and cancer type instances. For that, per model, we focused on the five most important features and identified instances with the top contributions to these features. An instance was considered a top contributor if its SHAP value for that feature that was among the 10% positive SHAP values (i.e., was a potential driver of the gain or loss) or the 10% negative SHAP values (i.e., was a potential driver of neutrality). The SHAP value for each instance and feature appears in Additional file 4 : Table S3. The list of instances and the features that they contributed to appears in Additional file 5 : Table S4. We then associated each instance with the number of features in which it was a top contributor. Next, we tested the impact of the strongest potential driver instances on the five most important features of the model. This was done by excluding from the dataset chromosome-arm and cancer type instances that were top contributors to at least three of the five features and repeating the construction and interpretation of each model using the revised dataset.
Correlation analysis
We correlated between feature values and the frequency of chromosome-arm gain or loss. The frequency of chromosome-arm gain/loss in cancers was obtained from GISTIC2.0 [ 24 ]. The frequency of chromosome-arm gain/loss in CCLs were obtained from [ 37 ]. Per chromosome-arm, its gain (loss) frequency was set to the median gain (loss) across cancers or CCLs. The feature value was set to median across cancers or CCLs. We used Spearman correlation, and p -values were adjusted using Benjamini–Hochberg procedure [ 71 ].
Paralog compensation analysis
For each cancer type and chromosome-arm, we considered all paralog pairs in which one of the genes resides on that chromosome-arm. We focused on recurrently lost genes per cancer type as defined by GISTIC2.0 [ 24 ]. We divided those genes by their minimal CRISPR essentiality score in CCLs that match the same cancer type (Additional file 2 : Table S1). Genes with a score ≤ − 0.5 were considered essential, and genes with a score ≥ − 0.3 were considered non-essential. Other genes were considered intermediate. Per gene, we checked whether its paralog was recurrently gained, lost, or neutral, in the same cancer, as detailed in Additional file 1 : Fig. S17A.
Chromosome-arm aneuploidy patterns in CCLs
Aneuploidy patterns were available for all (39) chromosome-arms in 14 CCLs from [ 37 ]. A chromosome-arm was considered as gained or lost in a CCL if the q -value of its amplification or deletion, respectively, was smaller than 0.15 (in case of ties, decision was made based on the lower q -value). In case of equal significant q -values, a chromosome-arm was considered as gained or lost based on their frequencies. Otherwise, the chromosome-arm was considered as neutral.
Construction of a feature dataset of instances of chromosome-arm and CCL pairs
The features dataset was similar to the dataset created for cancers, with the following exceptions. In features of cancer tissues, we replaced the transcriptomic features of cancers with transcriptomic features of CCLs. We obtained transcriptomic data of 25 CCLs from DepMap [ 27 ] and constructed the feature values per chromosome-arm and CCL as described above per chromosome-arm and cancer. Development features were removed since only a small number of CCLs had a matching organ. The final dataset contained features for all instances of 39 chromosome-arms and 10 CCLs for which the cancer’s normal tissue of origin was available in GTEx.
Cell culture
DLD1-WT cells and DLD1-Ts13 cells were cultured in RPMI-1640 (Life Technologies) with 10% fetal bovine serum (Sigma-Aldrich) and 1% penicillin–streptomycin-glutamine (Life Technologies). Cells were incubated at 37 °C with 5% CO2 and passaged twice a week using Trypsin–EDTA (0.25%) (Life Technologies). Cells were tested for mycoplasma contamination using the MycoAlert Mycoplasma Detection Kit (Lonza), according to the manufacturer’s instructions.
Cells were harvested using Bio-TRI® (Bio-Lab) and RNA was extracted following manufacturer’s protocol. cDNA was amplified using GoScript™ Reverse Transcription System (Promega) following manufacturer’s protocol. qRT-PCR was performed using Sybr® green, and quantification was performed using the ΔCT method. The following primer sequences were used: human KLF5 , forward, 5' ACACCAGACCGCAGCTCCA 3' and reverse 5' TCCATTGCTGCTGTCTGATTTGTAG 3', human NEK3 , forward, 5’ TACCCAAATGTGCCTTGGAG 3’, reverse 5’ ATCGGATTGGAGAGAAGACG 3’, human TTC7A , forward 5’ CTCGTGACCTGCAGACAAG 3’, reverse 5’ GGCTCCTAAAGTCTCCCAGC 3’.
siRNA transfection
For siRNA experiments, cells were plated in 96-well plates at 6000 cells per well and treated with compounds 24 h later. The cells were transfected with 15 nM siRNA against KLF5 (ONTARGETplus SMART-POOL®, Dharmacon) or with a control siRNA at the respective concentration (ONTARGETplus SMART-POOL®, Dharmacon) using Lipofectamine® RNAiMAX (Invitrogen) following the manufacturer’s protocol. Alternatively, for siRNA experiments against NEK3 and TTC7A , and for additional KLF5 experiments, cells were plated in 6-well plates at 400,000 cells per well and treated with compounds 24 h later. The cells were transfected with 30 nM against NEK3 and TTC7A or with 5 nM and 10 nM against KLF5 ; 48 h post seeding, the cells were split and plated in 96-wells at 10,000 cells per well. The effect of the knockdown against KLF5 , NEK3 , or TTC7A on cell viability/proliferation was measured by live cell imaging using Incucyte® (Satorius) or by the MTT assay (Sigma M2128) at 72 h (or at the indicated time point) post-transfection; 500 µg/mL MTT salt was diluted in complete medium and incubated at 37°C for 2 h. Formazan crystals were extracted using 10% Triton X-100 and 0.1 N HCl in isopropanol, and color absorption was quantified at 570 nm and 630 nm (Alliance Q9, Uvitec).
Cancer cell line and tumor data analysis
mRNA gene expression values, arm-level CNAs, CRISPR, and RNAi dependency scores (Chronos and DEMETER2 scores, respectively) were obtained from DepMap 22Q4 release ( www.depmap.org ). Effect size, p -values, and q -values (Fig. 4 A,C,E, Fig. 5 C) were taken directly from DepMap and were calculated as described in Tsherniak et al. TCGA mRNA gene expression values were obtained using the Xena browser [ 63 ]. Tumor arm-level alterations were retrieved from Taylor et al. 2018, Cancer Cell. Effect size, Spearman’s R and p -values in Fig. 4 G and Fig. 5 F were calculated using R functions. All colorectal cancer cell lines ( n = 85) and colorectal tumors ( n = 434) were included in the analyses.
The analyses that led to our choice of the paralog pair UCHL3 - UCHL1 are summarized in Additional file 6 : Table S5. In the left column are the paralogs that reside on chr-13q, which is frequently gained; in the adjacent column are the respective paralogs that reside on commonly lost chromosomes. The following columns describe the Spearman correlation between each paralog pair and the respective p -value. The right-hand columns describe the effect size of chr-13q paralogs’ gene expression between CRC cell lines with and without chr13q gain. Our criteria for finding appropriate paralog pairs for further analysis were as follows: firstly, to have a high expression of the chr-13q paralogs in CRC cell lines. Secondly, we aimed to reach a significant correlation between chr13q-residing genes and the essentiality of their paralogs.
Statistical analyses
Statistical analysis was performed using GraphPad PRISM® 9.1 software. Details of the statistical tests were reported in figure legends. Error bars represent SD. All experiments were performed in at least three biological replicates.
Availability of data and materials
The code for all the analyses is available on GitHub [ 72 ]. The datasets that were processed to build the dataset for the ML methods are available on Zenodo [ 73 ]. This includes features of normal tissues that were extracted from TRACE [ 74 ], TCGA expression data of the different cancer types that were obtained from Xena [ 75 ], and CRISPR and RNAi datasets that were obtained from DepMap [ 76 ].
Ben-David U, Amon A. Context is everything: aneuploidy in cancer. Nat Rev Genet. 2020;21(1):44–62.
Article CAS PubMed Google Scholar
Shukla A, Nguyen THM, Moka SB, Ellis JJ, Grady JP, Oey H, et al. Chromosome arm aneuploidies shape tumour evolution and drug response. Nat Commun. 2020;11(1):449.
Article CAS PubMed PubMed Central Google Scholar
Vasudevan A, Baruah PS, Smith JC, Wang Z, Sayles NM, Andrews P, et al. Single-Chromosomal gains can function as metastasis suppressors and promoters in colon cancer. Dev Cell. 2020;52(4):413–28 e6.
Ben-David U, Ha G, Tseng YY, Greenwald NF, Oh C, Shih J, et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet. 2017;49(11):1567–75.
Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell. 2018;33(4):676–89 e3.
Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155(4):948–62.
Sack LM, Davoli T, Li MZ, Li Y, Xu Q, Naxerova K, et al. Profound tissue specificity in proliferation control underlies cancer drivers and aneuploidy patterns. Cell. 2018;173(2):499–514 e23.
Patkar S, Heselmeyer-Haddad K, Auslander N, Hirsch D, Camps J, Bronder D, et al. Hard wiring of normal tissue-specific chromosome-wide gene expression levels is an additional factor driving cancer type-specific aneuploidies. Genome Med. 2021;13(1):93.
Liu Y, Chen C, Xu Z, Scuoppo C, Rillahan CD, Gao J, et al. Deletions linked to TP53 loss drive cancer through p53-independent mechanisms. Nature. 2016;531(7595):471–5.
Zhou XP, Li YJ, Hoang-Xuan K, Laurent-Puig P, Mokhtari K, Longy M, et al. Mutational analysis of the PTEN gene in gliomas: molecular and pathological correlations. Int J Cancer. 1999;84(2):150–4.
Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110.
Alfieri F, Caravagna G, Schaefer MH. Cancer genomes tolerate deleterious coding mutations through somatic copy number amplifications of wild-type regions. Nat Commun. 2023;14(1):3594.
Sheltzer JM, Amon A. The aneuploidy paradox: costs and benefits of an incorrect karyotype. Trends Genet. 2011;27(11):446–53.
Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, et al. Universal patterns of selection in cancer and somatic tissues. Cell. 2017;171(5):1029–41 e21.
Shih J, Sarmashghi S, Zhakula-Kostadinova N, Zhang S, Georgis Y, Hoyt SH, et al. Cancer aneuploidies are shaped primarily by effects on tumour fitness. Nature. 2023;619(7971):793–800.
Zitnik M, Nguyen F, Wang B, Leskovec J, Goldenberg A, Hoffman MM. Machine learning for integrating data in biology and medicine: principles, practice, and opportunities. Inf Fusion. 2019;50:71–91.
Article PubMed Google Scholar
Han Y, Yang J, Qian X, Cheng WC, Liu SH, Hua X, et al. DriverML: a machine learning algorithm for identifying driver genes in cancer sequencing studies. Nucleic Acids Res. 2019;47(8): e45.
Luo P, Ding Y, Lei X, Wu FX. deepDriver: predicting cancer driver genes based on somatic mutations using deep convolutional neural networks. Front Genet. 2019;10:13.
Mostavi M, Chiu YC, Chen Y, Huang Y. CancerSiamese: one-shot learning for predicting primary and metastatic tumor types unseen during model training. BMC Bioinformatics. 2021;22(1):244.
Article PubMed PubMed Central Google Scholar
Ramirez R, Chiu YC, Hererra A, Mostavi M, Ramirez J, Chen Y, et al. Classification of cancer types using graph convolutional neural networks. Front Phys. 2020;8:203.
Chiu Y-C, Zheng S, Wang L-J, Iskra BS, Rao MK, Houghton PJ, et al. Predicting and characterizing a cancer dependency map of tumors with deep learning. Science Advances. 2021;7(34):eabh1275.
Lundberg SM, Lee S-I. A unified approach to interpreting model predictions. Adv Neural Inf Process Syst. 2017;30(9):4768–77.
Google Scholar
Rodriguez-Perez R, Bajorath J. Interpretation of compound activity predictions from complex machine learning models using local approximations and Shapley values. J Med Chem. 2020;63(16):8761–77.
Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41.
GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369(6509):1318–30.
Article Google Scholar
Nichols CA, Gibson WJ, Brown MS, Kosmicki JA, Busanovich JP, Wei H, et al. Loss of heterozygosity of essential genes represents a widespread class of potential cancer vulnerabilities. Nat Commun. 2020;11(1):2517.
Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a cancer dependency map. Cell. 2017;170(3):564–76 e16.
Basha O, Argov CM, Artzy R, Zoabi Y, Hekselman I, Alfandari L, et al. Differential network analysis of multiple human tissue interactomes highlights tissue-selective processes and genetic disorder genes. Bioinformatics. 2020;36(9):2821–8.
Greene CS, Krishnan A, Wong AK, Ricciotti E, Zelaya RA, Himmelstein DS, et al. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet. 2015;47(6):569–76.
Sharon M, Vinogradov E, Argov CM, Lazarescu O, Zoabi Y, Hekselman I, et al. The differential activity of biological processes in tissues and cell subsets can illuminate disease-related processes and cell-type identities. Bioinformatics. 2022;38(6):1584–92.
Barshir R, Hekselman I, Shemesh N, Sharon M, Novack L, Yeger-Lotem E. Role of duplicate genes in determining the tissue-selectivity of hereditary diseases. PLoS Genet. 2018;14(5): e1007327.
Jubran J, Hekselman I, Novack L, Yeger-Lotem E. Dosage-sensitive molecular mechanisms are associated with the tissue-specificity of traits and diseases. Comput Struct Biotechnol J. 2020;18:4024–32.
Kingsford C, Salzberg SL. What are decision trees? Nat Biotechnol. 2008;26(9):1011–3.
Kotsiantis SB. Decision trees: a recent overview. Artif Intell Rev. 2013;39:261–83.
McFarland JM, Ho ZV, Kugener G, Dempster JM, Montgomery PG, Bryan JG, et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat Commun. 2018;9(1):4610.
Cohen-Sharir Y, McFarland JM, Abdusamad M, Marquis C, Bernhard SV, Kazachkova M, et al. Aneuploidy renders cancer cells vulnerable to mitotic checkpoint inhibition. Nature. 2021;590(7846):486–91.
Prasad K, Bloomfield M, Levi H, Keuper K, Bernhard SV, Baudoin NC, et al. Whole-genome duplication shapes the aneuploidy landscape of human cancers. Cancer Res. 2022;82(9):1736–52.
Ben-David U, Siranosian B, Ha G, Tang H, Oren Y, Hinohara K, et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature. 2018;560(7718):325–30.
Dempster JM, Boyle I, Vazquez F, Root DE, Boehm JS, Hahn WC, et al. Chronos: a cell population dynamics model of CRISPR experiments that improves inference of gene fitness effects. Genome Biol. 2021;22(1):343.
Chen C, Bhalala HV, Qiao H, Dong JT. A possible tumor suppressor role of the KLF5 transcription factor in human breast cancer. Oncogene. 2002;21(43):6567–72.
Ma J-B, Bai J-Y, Zhang H-B, Jia J, Shi Q, Yang C, et al. KLF5 inhibits STAT3 activity and tumor metastasis in prostate cancer by suppressing IGF1 transcription cooperatively with HDAC1. Cell Death Dis. 2020;11(6):466.
Luo Y, Chen C. The roles and regulation of the KLF5 transcription factor in cancers. Cancer Sci. 2021;112(6):2097–117.
McConnell BB, Bialkowska AB, Nandan MO, Ghaleb AM, Gordon FJ, Yang VW. Haploinsufficiency of Kruppel-like factor 5 rescues the tumor-initiating effect of the Apc(Min) mutation in the intestine. Cancer Res. 2009;69(10):4125–33.
Rutledge SD, Douglas TA, Nicholson JM, Vila-Casadesus M, Kantzler CL, Wangsa D, et al. Selective advantage of trisomic human cells cultured in non-standard conditions. Sci Rep. 2016;6:22828.
Chen WH, Zhao XM, van Noort V, Bork P. Human monogenic disease genes have frequently functionally redundant paralogs. PLoS Comput Biol. 2013;9(5): e1003073.
Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350(6264):1096–101.
Ito T, Young MJ, Li R, Jain S, Wernitznig A, Krill-Burger JM, et al. Paralog knockout profiling identifies DUSP4 and DUSP6 as a digenic dependence in MAPK pathway-driven cancers. Nat Genet. 2021;53(12):1664–72.
Zapata L, Pich O, Serrano L, Kondrashov FA, Ossowski S, Schaefer MH. Negative selection in tumor genome evolution acts on essential cellular functions and the immunopeptidome. Genome Biol. 2018;19(1):1–17.
de Kegel B, Ryan CJ. Paralog dispensability shapes homozygous deletion patterns in tumor genomes. Mol Syst Biol. 2023;19(12):e11987. https://doi.org/10.15252/msb.202311987 .
Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45(10):1134–40.
Cai Y, Crowther J, Pastor T, Abbasi Asbagh L, Baietti MF, De Troyer M, et al. Loss of chromosome 8p governs tumor progression and drug response by altering lipid metabolism. Cancer Cell. 2016;29(5):751–66.
Girish V, Lakhani AA, Thompson SL, Scaduto CM, Brown LM, Hagenson RA, et al. Oncogene-like addiction to aneuploidy in human cancers. Science. 2023;381(6660):eadg4521.
Zhao X, Cohen EEW, William WN Jr, Bianchi JJ, Abraham JP, Magee D, et al. Somatic 9p24.1 alterations in HPV(-) head and neck squamous cancer dictate immune microenvironment and anti-PD-1 checkpoint inhibitor activity. Proc Natl Acad Sci U S A. 2022;119(47):e2213835119.
Ben-David U, Ha G, Khadka P, Jin X, Wong B, Franke L, et al. The landscape of chromosomal aberrations in breast cancer mouse models reveals driver-specific routes to tumorigenesis. Nat Commun. 2016;7:12160.
Simonovsky E, Sharon M, Ziv M, Mauer O, Hekselman I, Jubran J, et al. Predicting molecular mechanisms of hereditary diseases by using their tissue-selective manifestation. Mol Syst Biol. 2023;19(8):e11407. https://doi.org/10.15252/msb.202211407 .
Hua J, Xiong Z, Lowey J, Suh E, Dougherty ER. Optimal number of features as a function of sample size for various classification rules. Bioinformatics. 2005;21(8):1509–15.
Bakker B, Taudt A, Belderbos ME, Porubsky D, Spierings DC, de Jong TV, et al. Single-cell sequencing reveals karyotype heterogeneity in murine and human malignancies. Genome Biol. 2016;17(1):115.
Gao R, Bai S, Henderson YC, Lin Y, Schalck A, Yan Y, et al. Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nat Biotechnol. 2021;39(5):599–608.
Gao R, Davis A, McDonald TO, Sei E, Shi X, Wang Y, et al. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat Genet. 2016;48(10):1119–30.
Gavish A, Tyler M, Greenwald AC, Hoefflin R, Simkin D, Tschernichovsky R, et al. Hallmarks of transcriptional intratumour heterogeneity across a thousand tumours. Nature. 2023;618(7965):598–606.
Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104(50):20007–12.
Center BITGDA. SNP6 copy number analysis (GISTIC2). Broad Institute of MIT and Harvard. 2016. https://gdac.broadinstitute.org/runs/analyses__latest/reports/cancer/STAD-TP/CopyNumber_Gistic2/nozzle.html .
Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–8.
Basha O, Flom D, Barshir R, Smoly I, Tirman S, Yeger-Lotem E. MyProteinNet: build up-to-date protein interaction networks for organisms, tissues and user-defined contexts. Nucleic Acids Res. 2015;43(W1):W258–63.
Sonawane AR, Platig J, Fagny M, Chen C-Y, Paulson JN, Lopes-Ramos CM, et al. Understanding tissue-specific gene regulation. Cell Rep. 2017;21(4):1077–88.
Cardoso-Moreira M, Halbert J, Valloton D, Velten B, Chen C, Shao Y, et al. Gene expression across mammalian organ development. Nature. 2019;571(7766):505–9.
Cunningham F, Allen JE, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, et al. Ensembl 2022. Nucleic Acids Res. 2022;50(D1):D988–95.
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in Python. The Journal of machine Learning research. 2011;12:2825–30.
Chen T, Guestrin C, editors. Xgboost: a scalable tree boosting system. Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining; 2016. p. 785–94. https://doi.org/10.1145/2939672.2939785 .
Lundberg SM, Erion G, Chen H, DeGrave A, Prutkin JM, Nair B, et al. From local explanations to global understanding with explainable AI for trees. Nat Mach Intell. 2020;2(1):56–67.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc: Ser B (Methodol). 1995;57(1):289–300.
Jubran J, Yeger-Lotem E. Machine-learning analysis of factors that shape cancer aneuploidy landscapes reveals an important role for negative selection. GitHub https://github.com/JumanJubran/AneuploidyML .
Jubran J, Yeger-Lotem E. Machine-learning analysis of factors that shape cancer aneuploidy landscapes reveals an important role for negative selection. Zenodo. https://zenodo.org/records/8199048 .
Simonovsky E, Yeger-Lotem E. Predicting molecular mechanisms of hereditary diseases by using their tissue-selective manifestation. Datasets. Zenodo. https://zenodo.org/records/10115922 .
Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Datasets. Xena. https://xenabrowser.net/datapages/?hub=https://gdc.xenahubs.net:443 .
Tsherniak A, Vazquez F, Montgomery P, Weir B, Kryukov G, Cowley G. Defining a cancer dependency map. Datasets. DepMap. https://depmap.org/portal/download/all/ .
Download references
Acknowledgements
The authors would like to thank Jason Sheltzer for providing DLD1-WT and DLD1 Ts13 cell lines.
J.J. wishes to thank the Baroness Ariane de Rothschild Women Doctoral Program.
Peer review information
Andrew Cosgrove was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Review history
The review history is available as Additional File 8 .
This study was funded by the Israel Science Foundation [401/22 to E.Y.-L.] and by a Ben-Gurion University grant [to E.Y.-L.]. Work in the Ben-David lab is supported by the European Research Council Starting Grant (grant #945674 to U.B.-D.), the Israel Science Foundation (grant #1805/21 to U.B.-D.), the Israel Cancer Research Fund (Project Grant to U.B.-D.), and the BSF Project Grant (grant #2019228 to U.B.-D.), and by the EMBO Young Investigator Program (to U.B.-D.).
Author information
Juman Jubran and Rachel Slutsky are equally contributing first authors.
Uri Ben-David and Esti Yeger-Lotem are equally contributing last authors.
Authors and Affiliations
Department of Clinical Biochemistry and Pharmacology, Ben-Gurion University of the Negev, 84105, Beer Sheva, Israel
Juman Jubran & Esti Yeger-Lotem
Department of Human Molecular Genetics and Biochemistry, Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel
Rachel Slutsky, Nir Rozenblum & Uri Ben-David
Department of Software & Information Systems Engineering, Ben-Gurion University of the Negev, 84105, Beer Sheva, Israel
Lior Rokach
The National Institute for Biotechnology in the Negev, Ben-Gurion University of the Negev, 84105, Beer Sheva, Israel
Esti Yeger-Lotem
You can also search for this author in PubMed Google Scholar
Contributions
U.B.-D. and E.Y.-L. conceived and oversaw the study. J.J. designed and performed the computational analyses and developed and interpreted the ML models. R.S. designed and performed the UCHL1 and KLF5 DepMap data analyses and the in vitro experiments. N.R. assisted with the in vitro experiments. L.R. advised on the ML analyses. J.J., R.S., U.B.-D., and E.Y.-L. analyzed and interpreted the data and wrote the manuscript. All authors reviewed and approved the manuscript.
Authors’ Twitter handles
Twitter handles: @yegerlotemlab (Esti Yeger-Lotem), @BenDavidLab (Uri Ben-David).
Corresponding authors
Correspondence to Uri Ben-David or Esti Yeger-Lotem .
Ethics declarations
Ethics approval and consent to participate.
Ethics approval is not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: supplementary figures..
This file contains Supplementary Figures S1-S17.
Additional file 2: Table S1.
Association of TCGA cancer types with normal tissues-of-origin and matching cell lines.
Additional file 3: Table S2.
The auROC and auPRC performance of ML models whose features were calculated using distinct percentages of genes.
Additional file 4: Table S3.
SHAP value per feature of each instance of chromosome-arm and tumor type in the gain and loss models.
Additional file 5: Table S4.
Potential driver instances of each feature in the gain and loss models, and their frequencies.
Additional file 6: Table S5.
Correlations between chr-13q residing genes and the essentiality of their paralogs.
Additional file 7: Table S6.
Co-incidence of arm-level events in the different cancer types, and their frequencies.
Additional file 8.
Rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Jubran, J., Slutsky, R., Rozenblum, N. et al. Machine-learning analysis reveals an important role for negative selection in shaping cancer aneuploidy landscapes. Genome Biol 25 , 95 (2024). https://doi.org/10.1186/s13059-024-03225-7
Download citation
Received : 05 July 2023
Accepted : 26 March 2024
Published : 15 April 2024
DOI : https://doi.org/10.1186/s13059-024-03225-7
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Genome Biology
ISSN: 1474-760X
- Submission enquiries: [email protected]
- General enquiries: [email protected]

IMAGES
VIDEO
COMMENTS
Find articles. with all of the words. with the exact phrase. with at least one of the words. without the words. where my words occur. anywhere in the article. in the title of the article. Return articles authored by. e.g., "PJ Hayes" or McCarthy. Return articles published in. e.g., J Biol Chem or Nature.
Broaden your research with images and primary sources Broaden your research with images and primary sources. Harness the power of visual materials—explore more than 3 million images now on JSTOR. Search for images Enhance your scholarly research with underground newspapers, magazines, and journals.
Review articles are another great way to find scholarly primary research articles. Review articles are not considered "primary research", but they pull together primary research articles on a topic, summarize and analyze them. In Google Scholar, click on Review Articles at the left of the search results screen. Ask your professor whether review ...
Primary research articles report on a single study. In the health sciences, primary research articles generally describe the following aspects of the study: The study's hypothesis or research question; The number of participants in the study, generally referred to as the "n"
Primary Research Articles. To conduct and publish an experiment or research study, an author or team of authors designs an experiment, gathers data, then analyzes the data and discusses the results of the experiment. A published experiment or research study will therefore look very different from other types of articles (newspaper stories ...
Discover a digital archive of scholarly articles, spanning centuries of scientific research. User Guide Learn how to find and read articles of interest to you. ... Journals deposit all NIH-funded articles as defined by the NIH Public Access Policy. 44 Selective Deposit Programs. Publisher deposits a subset of articles from a collection of journals.
Databases like CINAHL allow you to select Research Article to retrieve research articles in your search. Tip #3 - Sections of the Research Article to look for When reading an article, make sure to look inside the abstract (and the full text) and scan for sections contained in many primary research studies such as Introduction, Participants ...
Your use of JSTOR indicates your acceptance of the , the , and that you are 16 or older. JSTOR is a digital library of academic journals, books, and primary sources.
Click here to get help from a Polk State Librarian. This guide goes over how to find and analyze primary research articles in the sciences (e.g. nutrition, health sciences and nursing, biology, chemistry, physics, sociology, psychology). In addition, the guide explains how to tell the difference between a primary source and a secondary source ...
When you run a search, find a promising article in your results list and then look at the record for that item (usually by clicking on the title). The full database record for an item usually includes an abstract or summary--sometimes prepared by the journal or database, but often written by the author(s) themselves.
Primary research is any research that you conduct yourself. It can be as simple as a 2-question survey, or as in-depth as a years-long longitudinal study. The only key is that data must be collected firsthand by you. Primary research is often used to supplement or strengthen existing secondary research.
How to Identify Primary Research Articles. A primary research article reports on an empirical research study conducted by the authors. The goal of a primary research article is to present the result of original research that makes a new contribution to the body of knowledge. Characteristics: Almost always published in a peer-reviewed journal
To search for primary articles in Google Scholar, first, go to the main page and select 'Advanced Search'. In the Advanced Search window, check off the box that says 'Only show results from content I can access' and then select 'Include Patents'. Finally, click on 'Search'. This will filter out all secondary sources such as ...
3 Ways to Find Research Articles in PubMed. 1. Filter (Limit) to Article Type. Most citations in PubMed are for journal articles. However, you may limit your retrieval based on the type of material the article represents. Use the Filters on the Results page sidebar and look at the Article Types checklist which contains a list of frequently ...
A primary research article typically contains the following section headings: "Methods"/"Materials and Methods"/"Experimental Methods"(different journals title this section in different ways) "Results" "Discussion" If you skim the article, you should find additional evidence that an experiment was conducted by the authors themselves.
Research databases. You can search for scholarly sources online using databases and search engines like Google Scholar. These provide a range of search functions that can help you to find the most relevant sources. If you are searching for a specific article or book, include the title or the author's name. Alternatively, if you're just ...
Primary Sources are: Researchers reporting first-hand about their new research; Includes some Journal articles and some Books (monographs) In contrast, Secondary Sources summarize, analyze or report the work of other researchers. The most common type of journal article you will find in the sciences deals with primary research.These articles describe an original experiment or analysis that adds ...
Primary Research refers to research that was conducted by the author of the article you're reading. So if you're reading an article and in the methodology section the author refers to recruiting participants, identifying a control group, etc. you can be pretty sure the author has conducted the research themselves. When you're asked to find ...
This video explains what primary research articles are and demonstrates how to find them using the CU Library and the MEd library guide.
A primary research article will almost always contains statistics, numerical data presented in tables. Also, primary research articles are written in very formal, very technical language. Because primary research articles are written in technical language by professional researchers for experts like themselves, the articles can be very hard to ...
7 Steps to an Effective PubMed/Medline Searchand How to Find Primary Research Articles . 1. Access PubMed via the Library's web site in order to have access to the full text of the journals the Library subscribes to. 2. Break your search into component concepts and enter terms for each concept separately so that
This video shows how to search pubmed and locate a primary research article for a particular topic.
Discussion of why the 5 C's of historical thinking are important when viewing primary sources. The History Project at UC Irvine's 6 C's of Primary Source Analysis. Worksheet to aid in primary source analysis. Primary & Secondary Sources Infographic.
What Are Primary Sources? Primary sources are materials that were created during the time in question. They are the evidence of a particular time and place and moment. Secondary sources provide analysis of other materials, but primary sources are the raw and unfiltered data.
Background Aneuploidy, an abnormal number of chromosomes within a cell, is a hallmark of cancer. Patterns of aneuploidy differ across cancers, yet are similar in cancers affecting closely related tissues. The selection pressures underlying aneuploidy patterns are not fully understood, hindering our understanding of cancer development and progression. Results Here, we apply interpretable ...
Most Southern Baptists are conservative and Republican and have corresponding political priorities, but some, particularly among those in the pews, lean more to the center. Almost 3 in 5 Southern Baptist laity (58%) and 3 in 4 leaders (75%) identify with the Republican party. A quarter of churchgoers (26%) and 7% of leaders are Democrats, while ...